Welcome to the Recombinant Antibody Network
The Recombinant Antibody Network is a consortium of highly integrated technology centers at UCSF, the University of Chicago, and the University of Toronto, unified under a common goal to generate therapeutic grade recombinant antibodies at a proteome wide scale for biology and biomedicine.
Given that over half the human proteome is not annotated and that functional antibodies are not reliably available, a complete set of validated antibodies would greatly advance all areas of biology, including cancer therapy and infectious disease control. To undertake these challenges, RAN is systematically and comprehensively profiling families of protein targets using novel, modern high-throughput in vitro technology.
Latest Publications
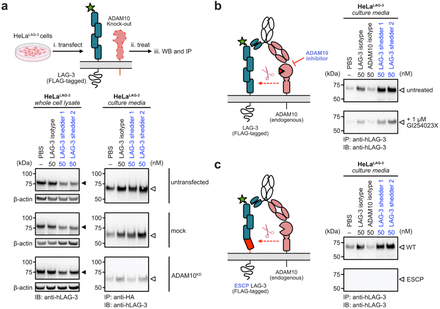
Yao Z; Zhao F; Miao K; Peters-Clarke T M; Zhang Y; Ganjave S D; Vázquez-Maldonado A L; Wu Y; Kumru K; Jumaa H; Leung K K; Wells J A
Targeted shedding of extracellular membrane proteins by induced protease recruitment Journal Article
In: bioRxiv, 2026, ISSN: 2692-8205.
@article{pmid41756920,
title = {Targeted shedding of extracellular membrane proteins by induced protease recruitment},
author = {Zi Yao and Fangzhu Zhao and Kun Miao and Trenton M Peters-Clarke and Yun Zhang and Snehal D Ganjave and Angel L Vázquez-Maldonado and Yan Wu and Kaan Kumru and Hammam Jumaa and Kevin K Leung and James A Wells},
doi = {10.64898/2026.02.17.706468},
issn = {2692-8205},
year = {2026},
date = {2026-02-01},
urldate = {2026-02-01},
journal = {bioRxiv},
abstract = {Extracellular targeted protein degradation has emerged as a promising therapeutic modality to eliminate proteins of interest (POIs) at the cell surface, by using bifunctional molecules to recruit natural recycling receptors or membrane-bound E3 ligases that redirect POIs to the lysosome. Another natural mechanism involves extracellular proteases that cleave and shed extracellular domains. Here, we exploit this endogenous mechanism by engineering bispecific antibody , that recruit a classic sheddase ADAM10 to POIs, inducing selective ectodomain shedding. We first targeted the immune checkpoint receptor LAG-3 and observed robust depletion of surface LAG-3 accompanied by accumulation of soluble LAG-3 fragments in both engineered cell lines and primary human T cells. Using biochemical and imaging assays, we confirmed that this antibody-induced shedding is restricted to extracellular protease activity and occurs independently of lysosomal trafficking. Notably, induced shedding of LAG-3 on activated primary T cells partially alleviated inhibitory signaling and reinvigorated IFN secretion. We extended the scope of induced shedding by developing that recognize synthetic epitope-tags that enabling rapid assessment of substrate compatibility across diverse targets. Using this platform, we identified multiple immune modulatory cell-surface receptors, including IL6Rα, CD62L and MIC-A that can be targeted for shedding. In summary, this work establishes a new paradigm for targeted extracellular proteolysis and expands the toolkit for studying extracellular proteolysis with potential therapeutic benefit.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
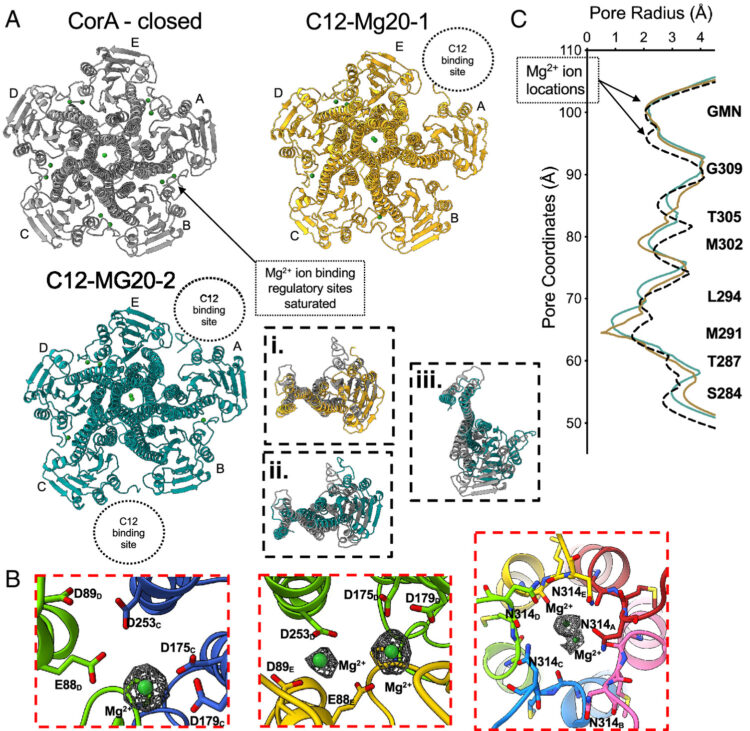
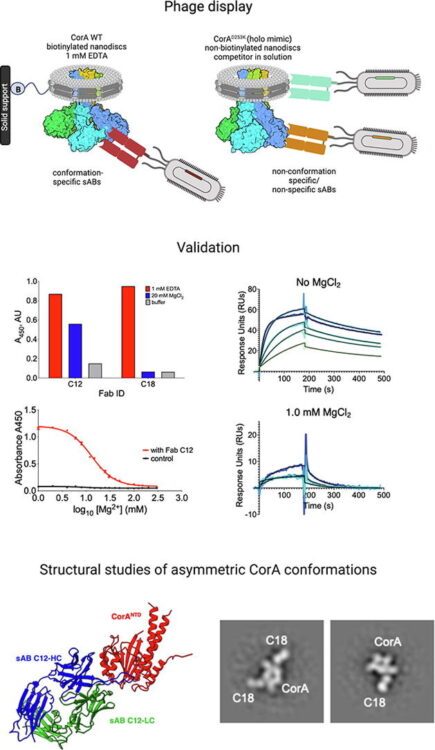
Erramilli S K; Nosol K; Pietrzak-Lichwa K; Schmandt N; Li T; Tokarz P; Hou J; Zhao M; Perozo E; Kossiakoff A A
Conformational ensembles of the magnesium channel CorA reveal structural basis for channel gating Journal Article
In: Proc Natl Acad Sci U S A, vol. 123, no. 8, pp. e2512532123, 2026, ISSN: 1091-6490.
@article{pmid41701836,
title = {Conformational ensembles of the magnesium channel CorA reveal structural basis for channel gating},
author = {Satchal K Erramilli and Kamil Nosol and Krzysztof Pietrzak-Lichwa and Nicolaus Schmandt and Tian Li and Piotr Tokarz and Jingkai Hou and Minglei Zhao and Eduardo Perozo and Anthony A Kossiakoff},
doi = {10.1073/pnas.2512532123},
issn = {1091-6490},
year = {2026},
date = {2026-02-01},
urldate = {2026-02-01},
journal = {Proc Natl Acad Sci U S A},
volume = {123},
number = {8},
pages = {e2512532123},
abstract = {In prokaryotes, CorA is the primary influx pathway for magnesium, a critical divalent cation in cellular physiology and biochemistry. Mechanistic studies show that homopentameric CorA is regulated through an intracellular [Mg]-dependent negative feedback loop, involving the asymmetric participation of individual subunits. To understand the connection between asymmetry and activation, we used single-particle cryo-EM to solve sixteen structures of nanodisc-reconstituted CorA. We utilized conformation-specific synthetic antibodies to stabilize subtle but significant conformational differences in the cryo-EM structures. Our results demonstrate that CorA exists as a set of conformational ensembles, where population size inversely correlates with intracellular Mg concentration. These ensembles include channels with a variety of pore conformations, both constricted and dilated, suggesting a spectrum of active CorA functional states. The ensembles connect asymmetric structural transitions in the cytoplasmic domain with conformational changes in the permeation pathway via an electrostatic network, ultimately controlling channel-gating events. We believe that these results establish a framework for understanding magnesium homeostasis in prokaryotic systems.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
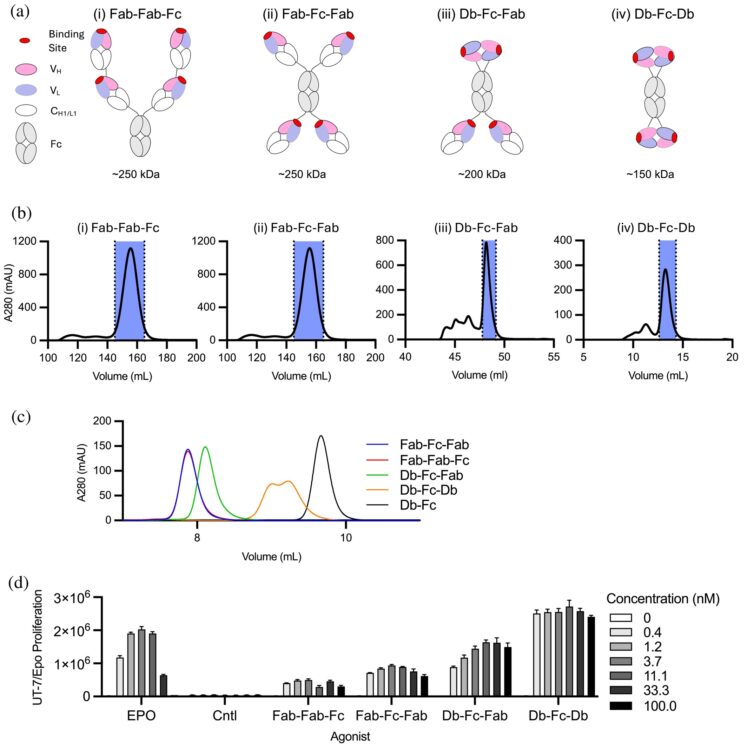
Adams J J; Blazer L L; Chung J; Karimi M; Davidson T; Blair B; Waddle C; Hokanson C A; Bruce H A; Singer A U; Tombak E; Gildemann K; Tamberg N; Kiiver K; Ustav M; Ma Y; Colombo L; Huang L J; Michnick S W; Moe O W; Sidhu S S
Tetravalent antibodies are more potent and efficacious erythropoiesis-stimulating agents than erythropoietin in vivo Journal Article
In: Protein Sci, vol. 35, no. 2, pp. e70462, 2026, ISSN: 1469-896X.
@article{pmid41556618,
title = {Tetravalent antibodies are more potent and efficacious erythropoiesis-stimulating agents than erythropoietin in vivo},
author = {Jarrett J Adams and Levi L Blazer and Jacky Chung and Minoo Karimi and Taylor Davidson and Bailey Blair and Carlos Waddle and Craig A Hokanson and Heather A Bruce and Alexander U Singer and Eva-Maria Tombak and Kiira Gildemann and Nele Tamberg and Kaja Kiiver and Mart Ustav and Yue Ma and Luigi Colombo and Lily Jun-Shen Huang and Stephen W Michnick and Orson W Moe and Sachdev S Sidhu},
doi = {10.1002/pro.70462},
issn = {1469-896X},
year = {2026},
date = {2026-02-01},
urldate = {2026-02-01},
journal = {Protein Sci},
volume = {35},
number = {2},
pages = {e70462},
abstract = {Recent studies have shown that tetravalent antibodies are potent and efficacious agonists of the erythropoietin (EPO) receptor (EPOR) both in vitro and in vivo. To identify antibody-based erythropoiesis-stimulating agents (ESAs) with therapeutic potential, we evaluated various tetravalent antibody formats for EPOR agonism and key biophysical properties necessary for biologic drug development. We identified two distinct tetravalent antibody formats that strongly stimulated the growth of UT7/Epo cells, which rely on EPOR signaling for proliferation. Moreover, one of these formats exhibited ideal biophysical characteristics for drug development. This format consisted of a diabody (Db) and two antigen-binding fragment (Fab) arms fused to the N- and C-termini of an Fc domain, respectively, to form a tetravalent Db-Fc-Fab (EPRA-0322). In a mouse model expressing the human EPOR, EPRA-0322 induced erythropoiesis with greater potency, efficacy, and duration than darbepoetin, a hyperglycosylated EPO currently used in clinical practice. These findings highlight tetravalent antibodies, and the Db-Fc-Fab format in particular, as promising next-generation ESAs suitable for large-scale production and clinical use.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Latest News
Recombinant Antibody Network Partners with Bristol Myers Squibb to Develop Novel Therapies
The Recombinant Antibody Network (RAN), a consortium comprising research groups from UC San Francisco, the…
Absolute Antibody Partners with the Recombinant Antibody Network to Facilitate Access to Engineered Recombinant Antibodies
Absolute Antibody Ltd., an industry-leading provider of recombinant antibody products and services, has announced a…
RAN to collaborate with Celgene on cancer therapeutics development
The RAN has recently signed a 3-year $25M agreement with the Celgene Corporation to develop next-generation,…
