Publications
View by year
- Publications: 2025
- Publications: 2024
- Publications: 2023
- Publications: 2022
- Publications: 2021
- Publications: 2020
- Publications: 2019
- Publications: 2018
- Publications: 2017
- Publications: 2016
- Publications: 2015
- Publications: 2014
- Publications: 2013
- Publications: 2012
Search
Search for author, keywords, or any other term.
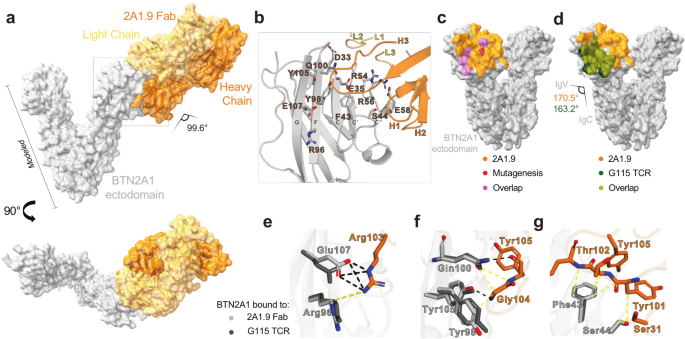
Ramesh, Amrita; Roy, Sobhan; Slezak, Tomasz; Fuller, James; Graves, Hortencia; Mamedov, Murad R.; Marson, Alexander; Kossiakoff, Anthony A.; Adams, Erin J.
Mapping the extracellular molecular architecture of the pAg-signaling complex with α-Butyrophilin antibodies Journal Article
In: Sci Rep, vol. 15, no. 1, 2025, ISSN: 2045-2322.
@article{Ramesh2025,
title = {Mapping the extracellular molecular architecture of the pAg-signaling complex with α-Butyrophilin antibodies},
author = {Amrita Ramesh and Sobhan Roy and Tomasz Slezak and James Fuller and Hortencia Graves and Murad R. Mamedov and Alexander Marson and Anthony A. Kossiakoff and Erin J. Adams},
doi = {10.1038/s41598-025-94347-w},
issn = {2045-2322},
year = {2025},
date = {2025-12-00},
urldate = {2025-12-00},
journal = {Sci Rep},
volume = {15},
number = {1},
publisher = {Springer Science and Business Media LLC},
abstract = {Target cells trigger Vγ9Vδ2 T cell activation by signaling the intracellular accumulation of phospho-antigen metabolites (pAgs) through Butyrophilin (BTN)-3A1 and BTN2A1 to the Vγ9Vδ2 T cell receptor (TCR). An incomplete understanding of the molecular dynamics in this signaling complex hampers Vγ9Vδ2 T cell immunotherapeutic efficacy. A panel of engineered α-BTN3A1 and α-BTN2A1 antibody (mAb) reagents was used to probe the roles of BTN3A1 and BTN2A1 in pAg signaling. Modified α-BTN3A1 mAbs with increased inter-Fab distances establish that tight clustering of BTN3A1 is not necessary to stimulate Vγ9Vδ2 T cell activation, and that antagonism may occur through occlusion of a critical binding interaction between BTN3A1 and a yet unknown co-receptor. Finally, a panel of additional α-BTN2A1 antagonists utilize different biophysical mechanisms to compete with Vγ9Vδ2 TCRs for BTN2A1 binding. The complex structures of BTN2A1 ectodomain and Fabs from three antagonist mAbs provide molecular insights into BTN2A1 epitopes critical for pAg-signaling.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
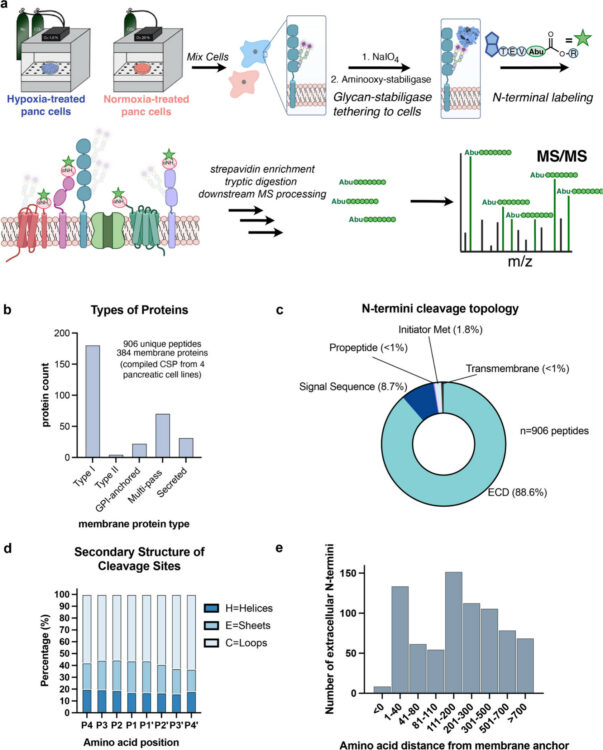
Lui, Irene; Schaefer, Kaitlin; Kirkemo, Lisa L; Zhou, Jie; Perera, Rushika M; Leung, Kevin K; Wells, James A
Hypoxia Induces Extensive Protein and Proteolytic Remodeling of the Cell Surface in Pancreatic Adenocarcinoma (PDAC) Cell Lines Journal Article
In: J Proteome Res, 2025, ISSN: 1535-3907.
@article{pmid40312771,
title = {Hypoxia Induces Extensive Protein and Proteolytic Remodeling of the Cell Surface in Pancreatic Adenocarcinoma (PDAC) Cell Lines},
author = {Irene Lui and Kaitlin Schaefer and Lisa L Kirkemo and Jie Zhou and Rushika M Perera and Kevin K Leung and James A Wells},
doi = {10.1021/acs.jproteome.4c01037},
issn = {1535-3907},
year = {2025},
date = {2025-05-01},
urldate = {2025-05-01},
journal = {J Proteome Res},
abstract = {The tumor microenvironment (TME) plays a crucial role in cancer progression. Hypoxia is a hallmark of the TME and induces a cascade of molecular events that affect cellular processes involved in metabolism, metastasis, and proteolysis. In pancreatic ductal adenocarcinoma (PDAC), tumor tissues are extremely hypoxic. Here, we leveraged mass spectrometry technologies to examine hypoxia-induced alterations in the abundance and proteolytic modifications to cell surface and secreted proteins. Across four PDAC cell lines, we discovered extensive proteolytic remodeling of cell surface proteins involved in cellular adhesion and motility. Looking outward at the surrounding secreted space, we identified hypoxia-regulated secreted and proteolytically shed proteins involved in regulating the humoral immune and inflammatory response, and an upregulation of proteins involved in metabolic processing and tissue development. Combining cell surface N-terminomics and secretomics to evaluate the cellular response to hypoxia enabled us to identify significantly altered candidate proteins which may serve as potential biomarkers and therapeutic targets in PDAC. Furthermore, this approach provides a blueprint for studying dysregulated extracellular proteolysis in other cancers and inflammatory diseases.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
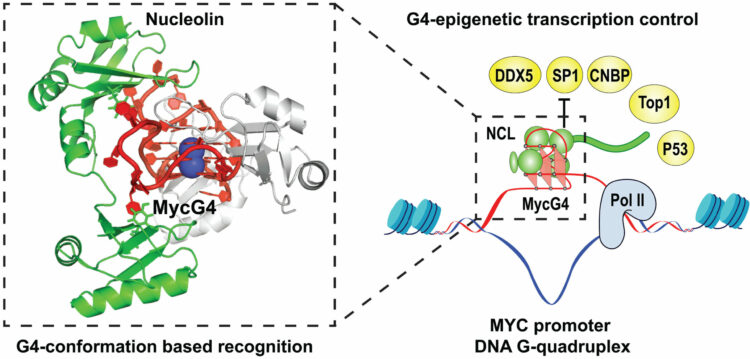
Chen, Luying; Dickerhoff, Jonathan; Zheng, Ke-wei; Erramilli, Satchal; Feng, Hanqiao; Wu, Guanhui; Onel, Buket; Chen, Yuwei; Wang, Kai-Bo; Carver, Megan; Lin, Clement; Sakai, Saburo; Wan, Jun; Vinson, Charles; Hurley, Laurence; Kossiakoff, Anthony A.; Deng, Nanjie; Bai, Yawen; Noinaj, Nicholas; Yang, Danzhou
Structural basis for nucleolin recognition of MYC promoter G-quadruplex Journal Article
In: Science, vol. 388, no. 6744, 2025, ISSN: 1095-9203.
@article{Chen2025,
title = {Structural basis for nucleolin recognition of MYC promoter G-quadruplex},
author = {Luying Chen and Jonathan Dickerhoff and Ke-wei Zheng and Satchal Erramilli and Hanqiao Feng and Guanhui Wu and Buket Onel and Yuwei Chen and Kai-Bo Wang and Megan Carver and Clement Lin and Saburo Sakai and Jun Wan and Charles Vinson and Laurence Hurley and Anthony A. Kossiakoff and Nanjie Deng and Yawen Bai and Nicholas Noinaj and Danzhou Yang},
doi = {10.1126/science.adr1752},
issn = {1095-9203},
year = {2025},
date = {2025-04-18},
urldate = {2025-04-18},
journal = {Science},
volume = {388},
number = {6744},
publisher = {American Association for the Advancement of Science (AAAS)},
abstract = {The MYC oncogene promoter G-quadruplex (MycG4) regulates transcription and is a prevalent G4 locus in immortal cells. Nucleolin, a major MycG4-binding protein, exhibits greater affinity for MycG4 than for nucleolin recognition element (NRE) RNA. Nucleolin’s four RNA binding domains (RBDs) are essential for high-affinity MycG4 binding. We present the 2.6-angstrom crystal structure of the nucleolin-MycG4 complex, revealing a folded parallel three-tetrad G-quadruplex with two coordinating potassium ions (K+), interacting with RBD1, RBD2, and Linker12 through its 6–nucleotide (nt) central loop and 5′ flanking region. RBD3 and RBD4 bind MycG4’s 1-nt loops as demonstrated by nuclear magnetic resonance (NMR). Cleavage under targets and tagmentation sequencing confirmed nucleolin’s binding to MycG4 in cells. Our results revealed a G4 conformation-based recognition by a regulating protein through multivalent interactions, suggesting that G4s are nucleolin’s primary cellular substrates, indicating G4 epigenetic transcriptional regulation and helping G4-targeted drug discovery.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Leung, Kevin K; Schaefer, Kaitlin; Lin, Zhi; Yao, Zi; Wells, James A
Engineered Proteins and Chemical Tools to Probe the Cell Surface Proteome Journal Article
In: Chem Rev, 2025, ISSN: 1520-6890.
@article{pmid40178992,
title = {Engineered Proteins and Chemical Tools to Probe the Cell Surface Proteome},
author = {Kevin K Leung and Kaitlin Schaefer and Zhi Lin and Zi Yao and James A Wells},
doi = {10.1021/acs.chemrev.4c00554},
issn = {1520-6890},
year = {2025},
date = {2025-04-01},
urldate = {2025-04-01},
journal = {Chem Rev},
abstract = {The cell surface proteome, or surfaceome, is the hub for cells to interact and communicate with the outside world. Many disease-associated changes are hard-wired within the surfaceome, yet approved drugs target less than 50 cell surface proteins. In the past decade, the proteomics community has made significant strides in developing new technologies tailored for studying the surfaceome in all its complexity. In this review, we first dive into the unique characteristics and functions of the surfaceome, emphasizing the necessity for specialized labeling, enrichment, and proteomic approaches. An overview of surfaceomics methods is provided, detailing techniques to measure changes in protein expression and how this leads to novel target discovery. Next, we highlight advances in proximity labeling proteomics (PLP), showcasing how various enzymatic and photoaffinity proximity labeling techniques can map protein-protein interactions and membrane protein complexes on the cell surface. We then review the role of extracellular post-translational modifications, focusing on cell surface glycosylation, proteolytic remodeling, and the secretome. Finally, we discuss methods for identifying tumor-specific peptide MHC complexes and how they have shaped therapeutic development. This emerging field of neo-protein epitopes is constantly evolving, where targets are identified at the proteome level and encompass defined disease-associated PTMs, complexes, and dysregulated cellular and tissue locations. Given the functional importance of the surfaceome for biology and therapy, we view surfaceomics as a critical piece of this quest for neo-epitope target discovery.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
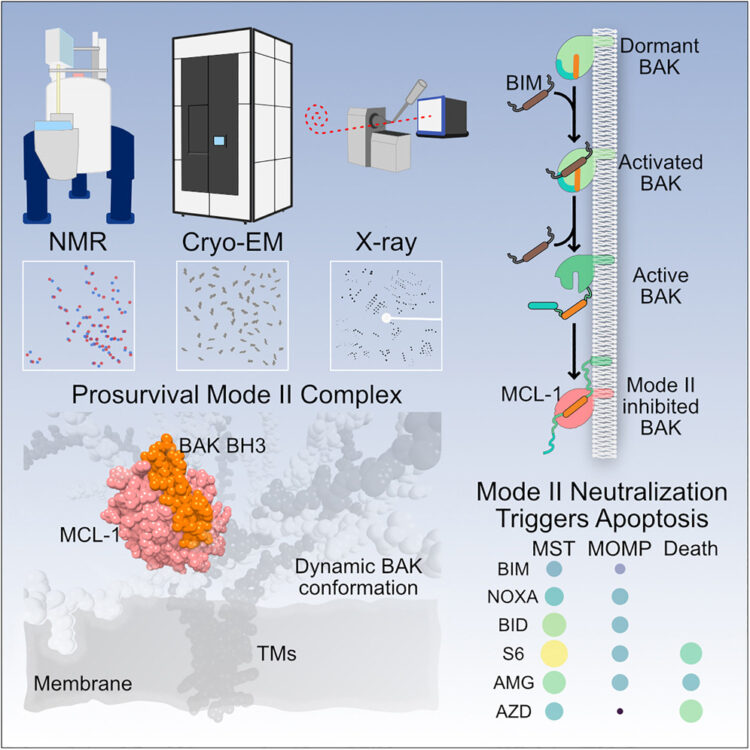
Srivastava, Shagun; Sekar, Giridhar; Ojoawo, Adedolapo; Aggarwal, Anup; Ferreira, Elisabeth; Uchikawa, Emiko; Yang, Meek; Grace, Christy R; Dey, Raja; Lin, Yi-Lun; Guibao, Cristina D; Jayaraman, Seetharaman; Mukherjee, Somnath; Kossiakoff, Anthony A; Dong, Bin; Myasnikov, Alexander; Moldoveanu, Tudor
Structural basis of BAK sequestration by MCL-1 in apoptosis Journal Article
In: Mol Cell, 2025, ISSN: 1097-4164.
@article{pmid40187349,
title = {Structural basis of BAK sequestration by MCL-1 in apoptosis},
author = {Shagun Srivastava and Giridhar Sekar and Adedolapo Ojoawo and Anup Aggarwal and Elisabeth Ferreira and Emiko Uchikawa and Meek Yang and Christy R Grace and Raja Dey and Yi-Lun Lin and Cristina D Guibao and Seetharaman Jayaraman and Somnath Mukherjee and Anthony A Kossiakoff and Bin Dong and Alexander Myasnikov and Tudor Moldoveanu},
doi = {10.1016/j.molcel.2025.03.013},
issn = {1097-4164},
year = {2025},
date = {2025-03-01},
urldate = {2025-03-01},
journal = {Mol Cell},
abstract = {Apoptosis controls cell fate, ensuring tissue homeostasis and promoting disease when dysregulated. The rate-limiting step in apoptosis is mitochondrial poration by the effector B cell lymphoma 2 (BCL-2) family proteins BAK and BAX, which are activated by initiator BCL-2 homology 3 (BH3)-only proteins (e.g., BIM) and inhibited by guardian BCL-2 family proteins (e.g., MCL-1). We integrated structural, biochemical, and pharmacological approaches to characterize the human prosurvival MCL-1:BAK complex assembled from their BCL-2 globular core domains. We reveal a canonical interaction with BAK BH3 bound to the hydrophobic groove of MCL-1 and disordered and highly dynamic BAK regions outside the complex interface. We predict similar conformations of activated effectors in complex with other guardians or effectors. The MCL-1:BAK complex is a major cancer drug target. We show that MCL-1 inhibitors are inefficient in neutralizing the MCL-1:BAK complex, requiring high doses to initiate apoptosis. Our study underscores the need to design superior clinical candidate MCL-1 inhibitors.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
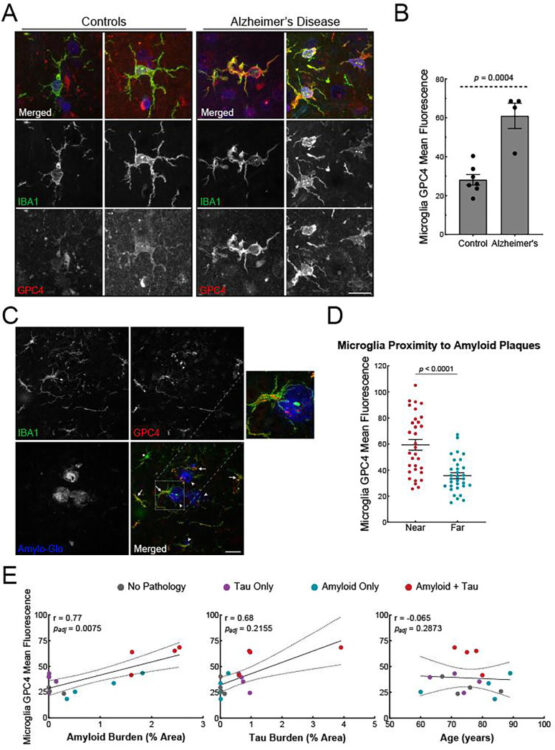
Holmes, Brandon B; Weigel, Thaddeus K; Chung, Jesseca M; Kaufman, Sarah K; Apresa, Brandon I; Byrnes, James R; Kumru, Kaan S; Vaquer-Alicea, Jaime; Gupta, Ankit; Rose, Indigo V L; Zhang, Yun; Nana, Alissa L; Alter, Dina; Grinberg, Lea T; Spina, Salvatore; Leung, Kevin K; Condello, Carlo; Kampmann, Martin; Seeley, William W; Coutinho-Budd, Jaeda C; Wells, James A
β-Amyloid Induces Microglial Expression of GPC4 and APOE Leading to Increased Neuronal Tau Pathology and Toxicity Journal Article
In: bioRxiv, 2025, ISSN: 2692-8205.
@article{pmid40060520,
title = {β-Amyloid Induces Microglial Expression of GPC4 and APOE Leading to Increased Neuronal Tau Pathology and Toxicity},
author = {Brandon B Holmes and Thaddeus K Weigel and Jesseca M Chung and Sarah K Kaufman and Brandon I Apresa and James R Byrnes and Kaan S Kumru and Jaime Vaquer-Alicea and Ankit Gupta and Indigo V L Rose and Yun Zhang and Alissa L Nana and Dina Alter and Lea T Grinberg and Salvatore Spina and Kevin K Leung and Carlo Condello and Martin Kampmann and William W Seeley and Jaeda C Coutinho-Budd and James A Wells},
doi = {10.1101/2025.02.20.637701},
issn = {2692-8205},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {bioRxiv},
abstract = {To elucidate the impact of Aβ pathology on microglia in Alzheimer's disease pathogenesis, we profiled the microglia surfaceome following treatment with Aβ fibrils. Our findings reveal that Aβ-associated human microglia upregulate Glypican 4 (GPC4), a GPI-anchored heparan sulfate proteoglycan (HSPG). In a amyloidosis model, glial GPC4 expression exacerbates motor deficits and reduces lifespan, indicating that glial GPC4 contributes to a toxic cellular program during neurodegeneration. In cell culture, GPC4 enhances microglia phagocytosis of tau aggregates, and shed GPC4 can act to facilitate tau aggregate uptake and seeding in neurons. Additionally, our data demonstrate that GPC4-mediated effects are amplified in the presence of APOE. These studies offer a mechanistic framework linking Aβ and tau pathology through microglial HSPGs and APOE.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Mann, Samuel I; Lin, Zhi; Tan, Sophia K; Zhu, Jiaqi; Widel, Zachary X W; Bakanas, Ian; Mansergh, Jarrett P; Liu, Rui; Kelly, Mark J S; Wu, Yibing; Wells, James A; Therien, Michael J; DeGrado, William F
De Novo Design of Proteins That Bind Naphthalenediimides, Powerful Photooxidants with Tunable Photophysical Properties Journal Article
In: J Am Chem Soc, 2025, ISSN: 1520-5126.
@article{pmid39982408,
title = {De Novo Design of Proteins That Bind Naphthalenediimides, Powerful Photooxidants with Tunable Photophysical Properties},
author = {Samuel I Mann and Zhi Lin and Sophia K Tan and Jiaqi Zhu and Zachary X W Widel and Ian Bakanas and Jarrett P Mansergh and Rui Liu and Mark J S Kelly and Yibing Wu and James A Wells and Michael J Therien and William F DeGrado},
doi = {10.1021/jacs.4c18151},
issn = {1520-5126},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {J Am Chem Soc},
abstract = { protein design provides a framework to test our understanding of protein function and build proteins with cofactors and functions not found in nature. Here, we report the design of proteins designed to bind powerful photooxidants and the evaluation of the use of these proteins to generate diffusible small-molecule reactive species. Because excited-state dynamics are influenced by the dynamics and hydration of a photooxidant's environment, it was important to not only design a binding site but also to evaluate its dynamic properties. Thus, we used computational design in conjunction with molecular dynamics (MD) simulations to design a protein, designated NBP (DI inding rotein), that held a naphthalenediimide (NDI), a powerful photooxidant, in a programmable molecular environment. Solution NMR confirmed the structure of the complex. We evaluated two NDI cofactors in this protein using ultrafast pump-probe spectroscopy to evaluate light-triggered intra- and intermolecular electron transfer function. Moreover, we demonstrated the utility of this platform to activate multiple molecular probes for protein labeling.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

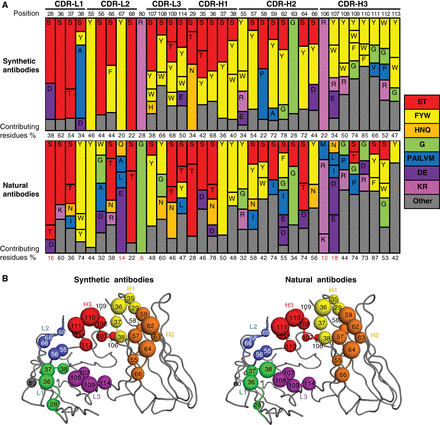
Gorelik, Maryna; Miersch, Shane; Sidhu, Sachdev S
Structural Survey of Antigen Recognition by Synthetic Human Antibodies Journal Article
In: Cold Spring Harb Protoc, vol. 2025, no. 2, pp. pdb.over107759, 2025, ISSN: 1559-6095.
@article{pmid38594044,
title = {Structural Survey of Antigen Recognition by Synthetic Human Antibodies},
author = {Maryna Gorelik and Shane Miersch and Sachdev S Sidhu},
doi = {10.1101/pdb.over107759},
issn = {1559-6095},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {Cold Spring Harb Protoc},
volume = {2025},
number = {2},
pages = {pdb.over107759},
abstract = {Synthetic antibody libraries have been used extensively to isolate and optimize antibodies. To generate these libraries, the immunological diversity and the antibody framework(s) that supports it outside of the binding regions are carefully designed/chosen to ensure favorable functional and biophysical properties. In particular, minimalist, single-framework synthetic libraries pioneered by our group have yielded a vast trove of antibodies to a broad array of antigens. Here, we review their systematic and iterative development to provide insights into the design principles that make them a powerful tool for drug discovery. In addition, the ongoing accumulation of crystal structures of antigen-binding fragment (Fab)-antigen complexes generated with synthetic antibodies enables a deepening understanding of the structural determinants of antigen recognition and usage of immunoglobulin sequence diversity, which can assist in developing new strategies for antibody and library optimization. Toward this, we also survey here the structural landscape of a comprehensive and unbiased set of 50 distinct complexes derived from these libraries and compare it to a similar set of natural antibodies with the goal of better understanding how each achieves molecular recognition and whether opportunities exist for iterative improvement of synthetic libraries. From this survey, we conclude that despite the minimalist strategies used for design of these synthetic antibody libraries, the overall structural interaction landscapes are highly similar to natural repertoires. We also found, however, some key differences that can help guide the iterative design of new synthetic libraries via the introduction of positionally tailored diversity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
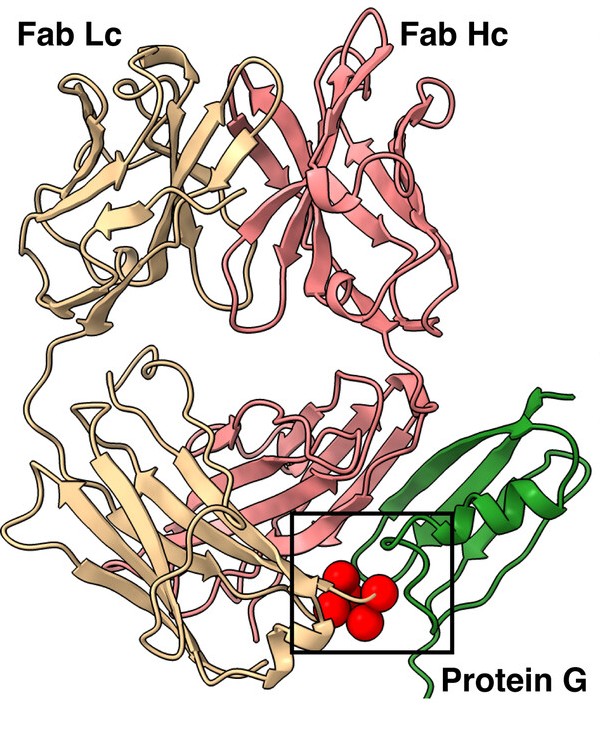
Slezak, Tomasz; O'Leary, Kelly M; Li, Jinyang; Rohaim, Ahmed; Davydova, Elena K; Kossiakoff, Anthony A
Engineered protein G variants for multifunctional antibody-based assemblies Journal Article
In: Protein Sci, vol. 34, no. 2, pp. e70019, 2025, ISSN: 1469-896X.
@article{pmid39865354,
title = {Engineered protein G variants for multifunctional antibody-based assemblies},
author = {Tomasz Slezak and Kelly M O'Leary and Jinyang Li and Ahmed Rohaim and Elena K Davydova and Anthony A Kossiakoff},
doi = {10.1002/pro.70019},
issn = {1469-896X},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {Protein Sci},
volume = {34},
number = {2},
pages = {e70019},
abstract = {We have developed a portfolio of antibody-based modules that can be prefabricated as standalone units and snapped together in plug-and-play fashion to create uniquely powerful multifunctional assemblies. The basic building blocks are derived from multiple pairs of native and modified Fab scaffolds and protein G (PG) variants engineered by phage display to introduce high pair-wise specificity. The variety of possible Fab-PG pairings provides a highly orthogonal system that can be exploited to perform challenging cell biology operations in a straightforward manner. The simplest manifestation allows multiplexed antigen detection using PG variants fused to fluorescently labeled SNAP-tags. Moreover, Fabs can be readily attached to a PG-Fc dimer module which acts as the core unit to produce plug-and-play IgG-like assemblies, and the utility can be further expanded to produce bispecific analogs using the "knobs into holes" strategy. These core PG-Fc dimer modules can be made and stored in bulk to produce off-the-shelf customized IgG entities in minutes, not days or weeks by just adding a Fab with the desired antigen specificity. In another application, the bispecific modalities form the building block for fabricating potent bispecific T-cell engagers (BiTEs), demonstrating their efficacy in cancer cell-killing assays. Additionally, the system can be adapted to include commercial antibodies as building blocks, greatly increasing the target space. Crystal structure analysis reveals that a few strategically positioned interactions engender the specificity between the Fab-PG variant pairs, requiring minimal changes to match the scaffolds for different possible combinations. This plug-and-play platform offers a user-friendly and versatile approach to enhance the functionality of antibody-based reagents in cell biology research.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
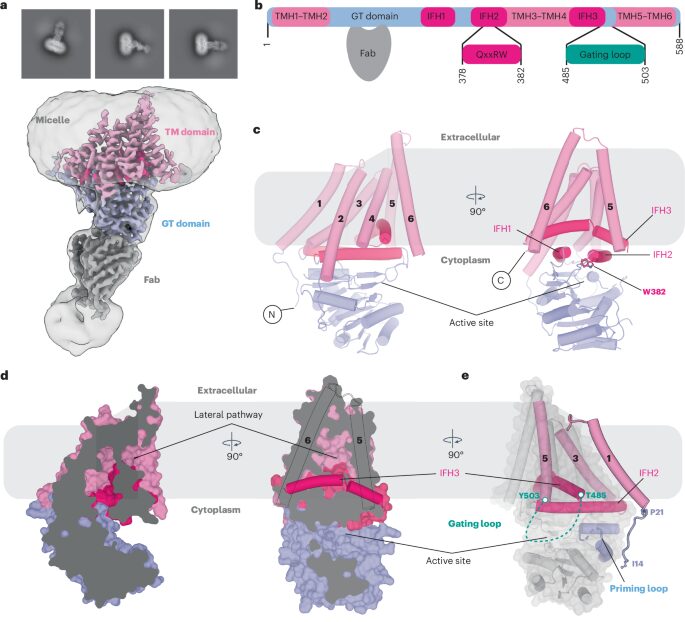
Górniak, Ireneusz; Stephens, Zachery; Erramilli, Satchal K; Gawda, Tomasz; Kossiakoff, Anthony A; Zimmer, Jochen
Structural insights into translocation and tailored synthesis of hyaluronan Journal Article
In: Nat Struct Mol Biol, vol. 32, no. 1, pp. 161–171, 2025, ISSN: 1545-9985.
@article{pmid39322765,
title = {Structural insights into translocation and tailored synthesis of hyaluronan},
author = {Ireneusz Górniak and Zachery Stephens and Satchal K Erramilli and Tomasz Gawda and Anthony A Kossiakoff and Jochen Zimmer},
doi = {10.1038/s41594-024-01389-1},
issn = {1545-9985},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Nat Struct Mol Biol},
volume = {32},
number = {1},
pages = {161--171},
abstract = {Hyaluronan (HA) is an essential component of the vertebrate extracellular matrix. It is a heteropolysaccharide of N-acetylglucosamine (GlcNAc) and glucuronic acid (GlcA) reaching several megadaltons in healthy tissues. HA is synthesized and translocated in a coupled reaction by HA synthase (HAS). Here, structural snapshots of HAS provide insights into HA biosynthesis, from substrate recognition to HA elongation and translocation. We monitor the extension of a GlcNAc primer with GlcA, reveal the coordination of the uridine diphosphate product by a conserved gating loop and capture the opening of a translocation channel to coordinate a translocating HA polymer. Furthermore, we identify channel-lining residues that modulate HA product lengths. Integrating structural and biochemical analyses suggests an avenue for polysaccharide engineering based on finely tuned enzymatic activity and HA coordination.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
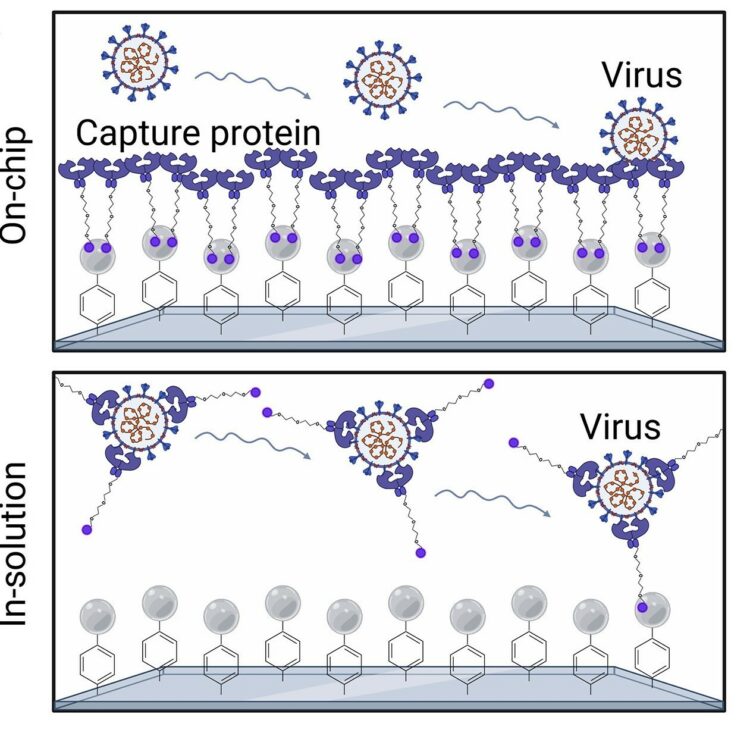
Rabe, Daniel C; Choudhury, Adarsh; Lee, Dasol; Luciani, Evelyn G; Ho, Uyen K; Clark, Alex E; Glasgow, Jeffrey E; Veiga, Sara; Michaud, William A; Capen, Diane; Flynn, Elizabeth A; Hartmann, Nicola; Garretson, Aaron F; Muzikansky, Alona; Goldberg, Marcia B; Kwon, Douglas S; Yu, Xu; Carlin, Aaron F; Theriault, Yves; Wells, James A; Lennerz, Jochen K; Lai, Peggy S; Rabi, Sayed Ali; Hoang, Anh N; Boland, Genevieve M; Stott, Shannon L
Ultrasensitive detection of intact SARS-CoV-2 particles in complex biofluids using microfluidic affinity capture Journal Article
In: Sci Adv, vol. 11, no. 2, pp. eadh1167, 2025, ISSN: 2375-2548.
@article{pmid39792670,
title = {Ultrasensitive detection of intact SARS-CoV-2 particles in complex biofluids using microfluidic affinity capture},
author = {Daniel C Rabe and Adarsh Choudhury and Dasol Lee and Evelyn G Luciani and Uyen K Ho and Alex E Clark and Jeffrey E Glasgow and Sara Veiga and William A Michaud and Diane Capen and Elizabeth A Flynn and Nicola Hartmann and Aaron F Garretson and Alona Muzikansky and Marcia B Goldberg and Douglas S Kwon and Xu Yu and Aaron F Carlin and Yves Theriault and James A Wells and Jochen K Lennerz and Peggy S Lai and Sayed Ali Rabi and Anh N Hoang and Genevieve M Boland and Shannon L Stott},
doi = {10.1126/sciadv.adh1167},
issn = {2375-2548},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Sci Adv},
volume = {11},
number = {2},
pages = {eadh1167},
abstract = {Measuring virus in biofluids is complicated by confounding biomolecules coisolated with viral nucleic acids. To address this, we developed an affinity-based microfluidic device for specific capture of intact severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Our approach used an engineered angiotensin-converting enzyme 2 to capture intact virus from plasma and other complex biofluids. Our device leverages a staggered herringbone pattern, nanoparticle surface coating, and processing conditions to achieve detection of as few as 3 viral copies per milliliter. We further validated our microfluidic assay on 103 plasma, 36 saliva, and 29 stool samples collected from unique patients with COVID-19, showing SARS-CoV-2 detection in 72% of plasma samples. Longitudinal monitoring in the plasma revealed our device's capacity for ultrasensitive detection of active viral infections over time. Our technology can be adapted to target other viruses using relevant cell entry molecules for affinity capture. This versatility underscores the potential for widespread application in viral load monitoring and disease management.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
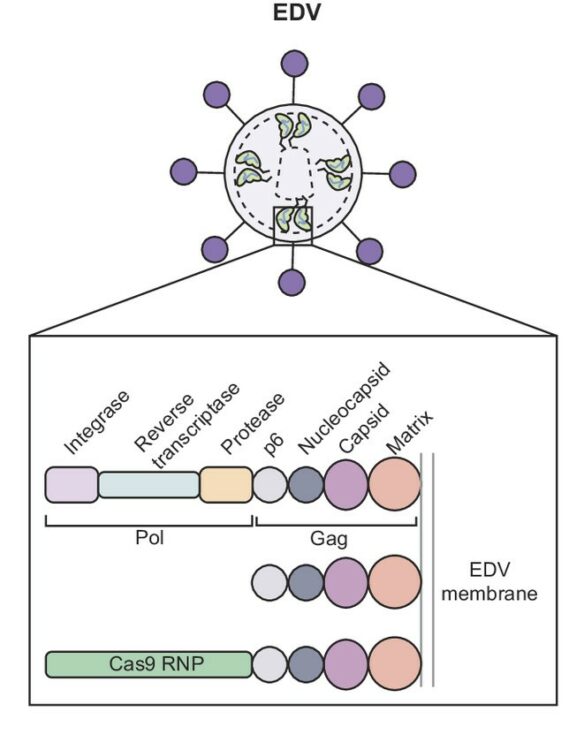
Ngo, Wayne; Peukes, Julia; Baldwin, Alisha; Xue, Zhiwei Wayne; Hwang, Sidney; Stickels, Robert R; Lin, Zhi; Satpathy, Ansuman T; Wells, James A; Schekman, Randy; Nogales, Eva; Doudna, Jennifer A
Mechanism-guided engineering of a minimal biological particle for genome editing Journal Article
In: Proc Natl Acad Sci U S A, vol. 122, no. 1, pp. e2413519121, 2025, ISSN: 1091-6490.
@article{pmid39793042,
title = {Mechanism-guided engineering of a minimal biological particle for genome editing},
author = {Wayne Ngo and Julia Peukes and Alisha Baldwin and Zhiwei Wayne Xue and Sidney Hwang and Robert R Stickels and Zhi Lin and Ansuman T Satpathy and James A Wells and Randy Schekman and Eva Nogales and Jennifer A Doudna},
doi = {10.1073/pnas.2413519121},
issn = {1091-6490},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Proc Natl Acad Sci U S A},
volume = {122},
number = {1},
pages = {e2413519121},
abstract = {The widespread application of genome editing to treat and cure disease requires the delivery of genome editors into the nucleus of target cells. Enveloped delivery vehicles (EDVs) are engineered virally derived particles capable of packaging and delivering CRISPR-Cas9 ribonucleoproteins (RNPs). However, the presence of lentiviral genome encapsulation and replication proteins in EDVs has obscured the underlying delivery mechanism and precluded particle optimization. Here, we show that Cas9 RNP nuclear delivery is independent of the native lentiviral capsid structure. Instead, EDV-mediated genome editing activity corresponds directly to the number of nuclear localization sequences on the Cas9 enzyme. EDV structural analysis using cryo-electron tomography and small molecule inhibitors guided the removal of ~80% of viral residues, creating a minimal EDV (miniEDV) that retains full RNP delivery capability. MiniEDVs are 25% smaller yet package equivalent amounts of Cas9 RNPs relative to the original EDVs and demonstrated increased editing in cell lines and therapeutically relevant primary human T cells. These results show that virally derived particles can be streamlined to create efficacious genome editing delivery vehicles with simpler production and manufacturing.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Kordon, Szymon P; Cechova, Kristina; Bandekar, Sumit J; Leon, Katherine; Dutka, Przemysław; Siffer, Gracie; Kossiakoff, Anthony A; Vafabakhsh, Reza; Araç, Demet
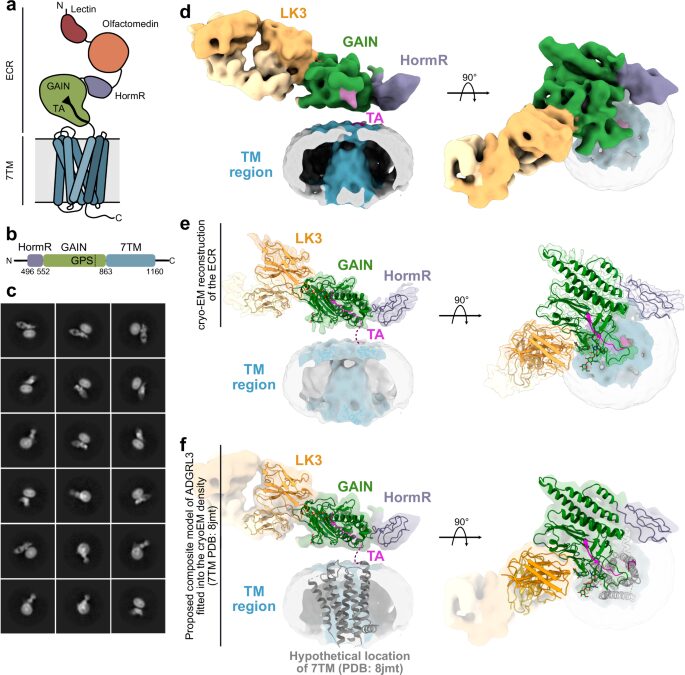
Conformational coupling between extracellular and transmembrane domains modulates holo-adhesion GPCR function Journal Article
In: Nat Commun, vol. 15, no. 1, pp. 10545, 2024, ISSN: 2041-1723.
@article{pmid39627215,
title = {Conformational coupling between extracellular and transmembrane domains modulates holo-adhesion GPCR function},
author = {Szymon P Kordon and Kristina Cechova and Sumit J Bandekar and Katherine Leon and Przemysław Dutka and Gracie Siffer and Anthony A Kossiakoff and Reza Vafabakhsh and Demet Araç},
doi = {10.1038/s41467-024-54836-4},
issn = {2041-1723},
year = {2024},
date = {2024-12-01},
urldate = {2024-12-01},
journal = {Nat Commun},
volume = {15},
number = {1},
pages = {10545},
abstract = {Adhesion G Protein-Coupled Receptors (aGPCRs) are key cell-adhesion molecules involved in numerous physiological functions. aGPCRs have large multi-domain extracellular regions (ECRs) containing a conserved GAIN domain that precedes their seven-pass transmembrane domain (7TM). Ligand binding and mechanical force applied on the ECR regulate receptor function. However, how the ECR communicates with the 7TM remains elusive, because the relative orientation and dynamics of the ECR and 7TM within a holoreceptor is unclear. Here, we describe the cryo-EM reconstruction of an aGPCR, Latrophilin3/ADGRL3, and reveal that the GAIN domain adopts a parallel orientation to the transmembrane region and has constrained movement. Single-molecule FRET experiments unveil three slow-exchanging FRET states of the ECR relative to the transmembrane region within the holoreceptor. GAIN-targeted antibodies, and cancer-associated mutations at the GAIN-7TM interface, alter FRET states, cryo-EM conformations, and receptor signaling. Altogether, this data demonstrates conformational and functional coupling between the ECR and 7TM, suggesting an ECR-mediated mechanism for aGPCR activation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
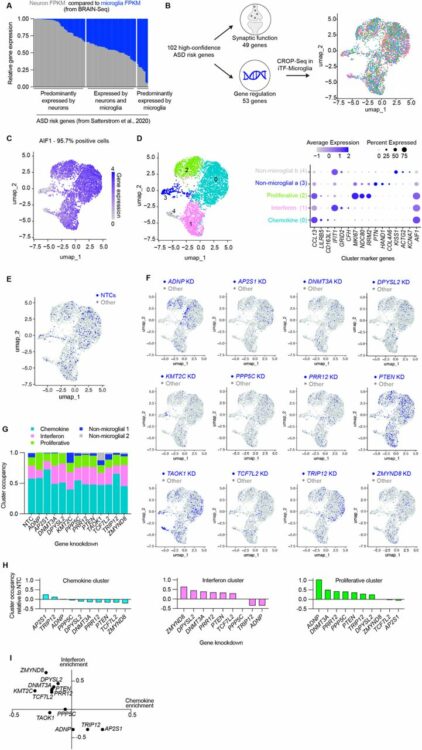
Teter, Olivia M; McQuade, Amanda; Hagan, Venus; Liang, Weiwei; Dräger, Nina M; Sattler, Sydney M; Holmes, Brandon B; Castillo, Vincent Cele; Papakis, Vasileios; Leng, Kun; Boggess, Steven; Nowakowski, Tomasz J; Wells, James; Kampmann, Martin
CRISPRi-based screen of Autism Spectrum Disorder risk genes in microglia uncovers roles of in microglia endocytosis and synaptic pruning Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39605704,
title = {CRISPRi-based screen of Autism Spectrum Disorder risk genes in microglia uncovers roles of in microglia endocytosis and synaptic pruning},
author = {Olivia M Teter and Amanda McQuade and Venus Hagan and Weiwei Liang and Nina M Dräger and Sydney M Sattler and Brandon B Holmes and Vincent Cele Castillo and Vasileios Papakis and Kun Leng and Steven Boggess and Tomasz J Nowakowski and James Wells and Martin Kampmann},
doi = {10.1101/2024.06.01.596962},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {Autism Spectrum Disorders (ASD) are a set of neurodevelopmental disorders with complex biology. The identification of ASD risk genes from exome-wide association studies and de novo variation analyses has enabled mechanistic investigations into how ASD-risk genes alter development. Most functional genomics studies have focused on the role of these genes in neurons and neural progenitor cells. However, roles for ASD risk genes in other cell types are largely uncharacterized. There is evidence from postmortem tissue that microglia, the resident immune cells of the brain, appear activated in ASD. Here, we used CRISPRi-based functional genomics to systematically assess the impact of ASD risk gene knockdown on microglia activation and phagocytosis. We developed an iPSC-derived microglia-neuron coculture system and high-throughput flow cytometry readout for synaptic pruning to enable parallel CRISPRi-based screening of phagocytosis of beads, synaptosomes, and synaptic pruning. Our screen identified , a high-confidence ASD risk genes, as a modifier of microglial synaptic pruning. We found that microglia with ADNP loss have altered endocytic trafficking, remodeled proteomes, and increased motility in coculture.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Martyn, Gregory D; Kalagiri, Rajasree; Veggiani, Gianluca; Stanfield, Robyn L; Choudhuri, Indrani; Sala, Margaux; Meisenhelder, Jill; Chen, Chao; Biswas, Avik; Levy, Ronald M; Lyumkis, Dmitry; Wilson, Ian A; Hunter, Tony; Sidhu, Sachdev S
Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39574610,
title = {Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody},
author = {Gregory D Martyn and Rajasree Kalagiri and Gianluca Veggiani and Robyn L Stanfield and Indrani Choudhuri and Margaux Sala and Jill Meisenhelder and Chao Chen and Avik Biswas and Ronald M Levy and Dmitry Lyumkis and Ian A Wilson and Tony Hunter and Sachdev S Sidhu},
doi = {10.1101/2024.11.04.621849},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {Histidine phosphorylation (pHis) is a non-canonical post-translational modification (PTM) that is historically understudied due to a lack of robust reagents that are required for its investigation, such as high affinity pHis-specific antibodies. Engineering pHis-specific antibodies is very challenging due to the labile nature of the phosphoramidate (P-N) bond and the stringent requirements for selective recognition of the two isoforms, 1-phosphohistidine (1-pHis) and 3-phosphohistidine (3-pHis). Here, we present a strategy for engineering of antibodies for detection of native 3-pHis targets. Specifically, we humanized the rabbit SC44-8 anti-3-pTza (a stable 3-pHis mimetic) mAb into a scaffold (herein referred to as hSC44) that was suitable for phage display. We then constructed six unique Fab phage-displayed libraries using the hSC44 scaffold and selected high affinity 3-pHis binders. Our selection strategy was carefully designed to enrich antibodies that bound 3-pHis with high affinity and had specificity for 3-pHis versus 3-pTza. hSC44.20N32F, the best engineered antibody, has an ~10-fold higher affinity for 3-pHis than the parental hSC44. Eleven new Fab structures, including the first reported antibody-pHis peptide structures were solved by X-ray crystallography. Structural and quantum mechanical calculations provided molecular insights into 3-pHis and 3-pTza discrimination by different hSC44 variants and their affinity increase obtained through engineering. Furthermore, we demonstrate the utility of these newly developed high-affinity 3-pHis-specific antibodies for recognition of pHis proteins in mammalian cells by immunoblotting and immunofluorescence staining. Overall, our work describes a general method for engineering PTM-specific antibodies and provides a set of novel antibodies for further investigations of the role of 3-pHis in cell biology.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Rathnayake, Sewwandi S; Erramilli, Satchal K; Kossiakoff, Anthony A; Vecchio, Alex J
Cryo-EM structures of Clostridium perfringens enterotoxin bound to its human receptor, claudin-4 Journal Article
In: Structure, vol. 32, no. 11, pp. 1936–1951.e5, 2024, ISSN: 1878-4186.
@article{pmid39383874,
title = {Cryo-EM structures of Clostridium perfringens enterotoxin bound to its human receptor, claudin-4},
author = {Sewwandi S Rathnayake and Satchal K Erramilli and Anthony A Kossiakoff and Alex J Vecchio},
doi = {10.1016/j.str.2024.09.015},
issn = {1878-4186},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {Structure},
volume = {32},
number = {11},
pages = {1936--1951.e5},
abstract = {Clostridium perfringens enterotoxin (CpE) causes prevalent and deadly gastrointestinal disorders. CpE binds to receptors called claudins on the apical surfaces of small intestinal epithelium. Claudins normally regulate paracellular transport but are hijacked from doing so by CpE and are instead led to form claudin/CpE complexes. Claudin/CpE complexes are the building blocks of oligomeric β-barrel pores that penetrate the plasma membrane and induce gut cytotoxicity. Here, we present the structures of CpE in complex with its native claudin receptor in humans, claudin-4, using cryogenic electron microscopy. The structures reveal the architecture of the claudin/CpE complex, the residues used in binding, the orientation of CpE relative to the membrane, and CpE-induced changes to claudin-4. Further, structures and modeling allude to the biophysical procession from claudin/CpE complexes to cytotoxic β-barrel pores during pathogenesis. In full, this work proposes a model of claudin/CpE assembly and provides strategies to obstruct its formation to treat CpE diseases.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
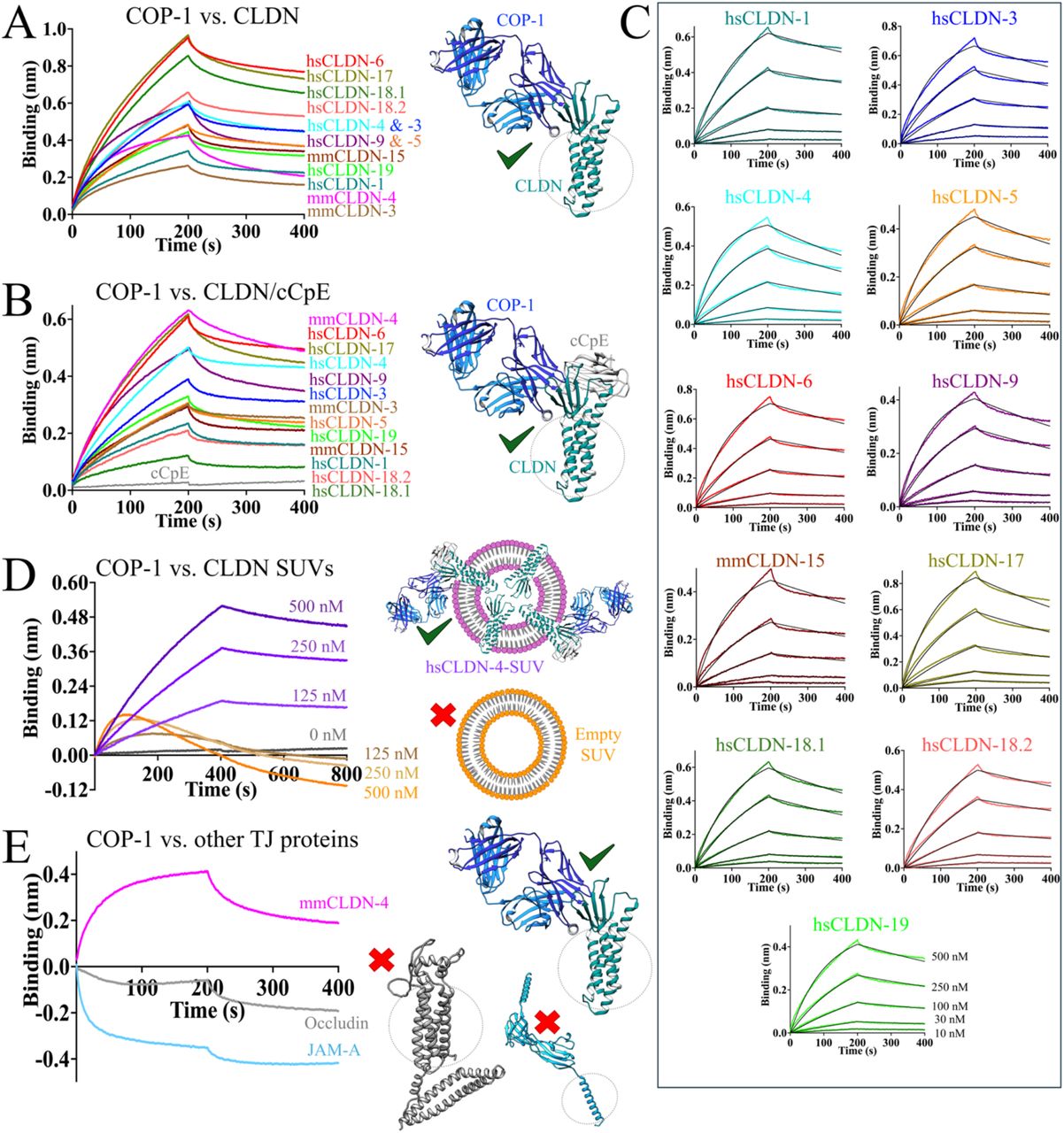
Ogbu, Chinemerem P; Mandriota, Alexandria M; Liu, Xiangdong; de Las Alas, Mason; Kapoor, Srajan; Choudhury, Jagrity; Kossiakoff, Anthony A; Duffey, Michael E; Vecchio, Alex J
Biophysical Basis of Paracellular Barrier Modulation by a Pan-Claudin-Binding Molecule Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39605593,
title = {Biophysical Basis of Paracellular Barrier Modulation by a Pan-Claudin-Binding Molecule},
author = {Chinemerem P Ogbu and Alexandria M Mandriota and Xiangdong Liu and Mason de Las Alas and Srajan Kapoor and Jagrity Choudhury and Anthony A Kossiakoff and Michael E Duffey and Alex J Vecchio},
doi = {10.1101/2024.11.10.622873},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {Claudins are a 27-member protein family that form and fortify specialized cell contacts in endothelium and epithelium called tight junctions. Tight junctions restrict paracellular transport across tissues by forming molecular barriers between cells. Claudin-binding molecules thus hold promise for modulating tight junction permeability to deliver drugs or as therapeutics to treat tight junction-linked disease. The development of claudin-binding molecules, however, is hindered by their intractability and small targetable surfaces. Here, we determine that a synthetic antibody fragment (sFab) we developed binds directly to 10 claudin subtypes with nanomolar affinity by targeting claudin's paracellular-exposed surface. Application of this sFab to cells that model intestinal epithelium show that it opens the paracellular barrier comparable to a known, but application limited, tight junction modulator. This novel pan-claudin-binding molecule can probe claudin or tight junction structure and holds potential as a broad modulator of tight junction permeability for basic or translational applications.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
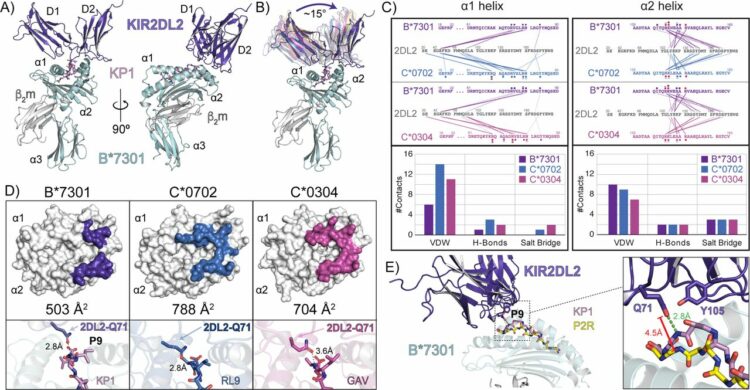
Ross, Philipp; Hilton, Hugo G; Lodwick, Jane; Slezak, Tomasz; Guethlein, Lisbeth A; McMurtrey, Curtis P; Han, Alex S; Nielsen, Morten; Yong, Daniel; Dulberger, Charles L; Nolan, Kristof T; Roy, Sobhan; Castro, Caitlin D; Hildebrand, William H; Zhao, Minglei; Kossiakoff, Anthony; Parham, Peter; Adams, Erin J
Molecular characterization of the archaic HLA-B*73:01 allele reveals presentation of a unique peptidome and skewed engagement by KIR2DL2 Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39651149,
title = {Molecular characterization of the archaic HLA-B*73:01 allele reveals presentation of a unique peptidome and skewed engagement by KIR2DL2},
author = {Philipp Ross and Hugo G Hilton and Jane Lodwick and Tomasz Slezak and Lisbeth A Guethlein and Curtis P McMurtrey and Alex S Han and Morten Nielsen and Daniel Yong and Charles L Dulberger and Kristof T Nolan and Sobhan Roy and Caitlin D Castro and William H Hildebrand and Minglei Zhao and Anthony Kossiakoff and Peter Parham and Erin J Adams},
doi = {10.1101/2024.11.25.625330},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {HLA class I alleles of archaic origin may have been retained in modern humans because they provide immunity against diseases to which archaic humans had evolved resistance. According to this model, archaic introgressed alleles were somehow distinct from those that evolved in African populations. Here we show that HLA-B*73:01, a rare allotype with putative archaic origins, has a relatively rare peptide binding motif with an unusually long-tailed peptide length distribution. We also find that HLA-B*73:01 combines a restricted and unique peptidome with high-cell surface expression, characteristics that make it well-suited to combat one or a number of closely-related pathogens. Furthermore, a crystal structure of HLA-B*73:01 in complex with KIR2DL2 highlights differences from previously solved structures with HLA-C molecules. These molecular characteristics distinguish HLA-B*73:01 from other HLA class I alleles previously investigated and may have provided early modern human migrants that inherited this allele with a selective advantage as they colonized Europe and Asia.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
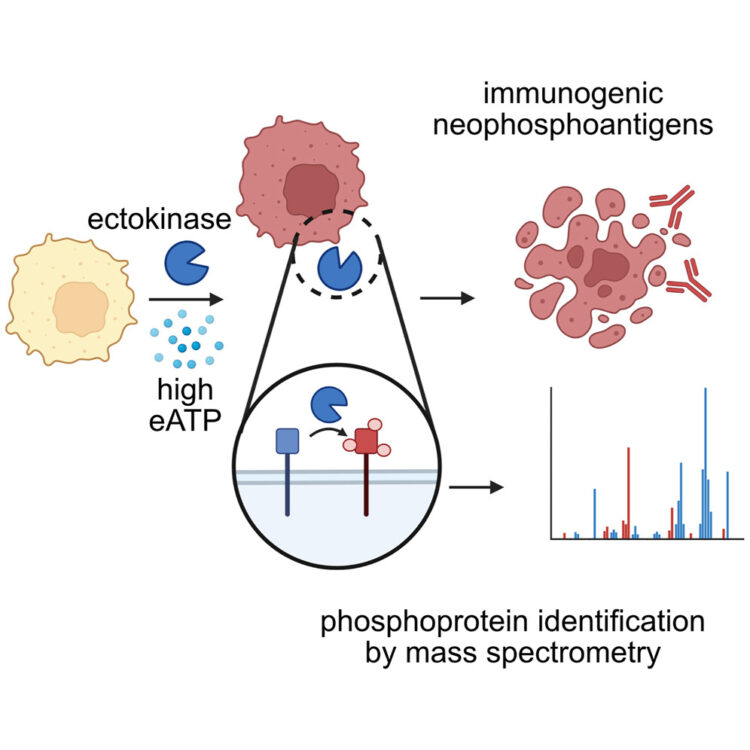
Delaveris, Corleone S; Kong, Sophie; Glasgow, Jeff; Loudermilk, Rita P; Kirkemo, Lisa L; Zhao, Fangzhu; Salangsang, Fernando; Phojanakong, Paul; Serrano, Juan Antonio Camara; Steri, Veronica; Wells, James A
Chemoproteomics reveals immunogenic and tumor-associated cell surface substrates of ectokinase CK2α Journal Article
In: Cell Chem Biol, vol. 31, no. 9, pp. 1729–1739.e9, 2024, ISSN: 2451-9448.
@article{pmid39178841,
title = {Chemoproteomics reveals immunogenic and tumor-associated cell surface substrates of ectokinase CK2α},
author = {Corleone S Delaveris and Sophie Kong and Jeff Glasgow and Rita P Loudermilk and Lisa L Kirkemo and Fangzhu Zhao and Fernando Salangsang and Paul Phojanakong and Juan Antonio Camara Serrano and Veronica Steri and James A Wells},
doi = {10.1016/j.chembiol.2024.07.018},
issn = {2451-9448},
year = {2024},
date = {2024-09-01},
urldate = {2024-09-01},
journal = {Cell Chem Biol},
volume = {31},
number = {9},
pages = {1729--1739.e9},
abstract = {Foreign epitopes for immune recognition provide the basis of anticancer immunity. Due to the high concentration of extracellular adenosine triphosphate in the tumor microenvironment, we hypothesized that extracellular kinases (ectokinases) could have dysregulated activity and introduce aberrant phosphorylation sites on cell surface proteins. We engineered a cell-tethered version of the extracellular kinase CK2α, demonstrated it was active on cells under tumor-relevant conditions, and profiled its substrate scope using a chemoproteomic workflow. We then demonstrated that mice developed polyreactive antisera in response to syngeneic tumor cells that had been subjected to surface hyperphosphorylation with CK2α. Interestingly, these mice developed B cell and CD4 T cell responses in response to these antigens but failed to develop a CD8 T cell response. This work provides a workflow for probing the extracellular phosphoproteome and demonstrates that extracellular phosphoproteins are immunogenic even in a syngeneic system.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Samassekou, Kadidia; Garland-Kuntz, Elisabeth E; Ohri, Vaani; Fisher, Isaac J; Erramilli, Satchal K; Muralidharan, Kaushik; Bogdan, Livia M; Gick, Abigail M; Kossiakoff, Anthony A; Lyon, Angeline M
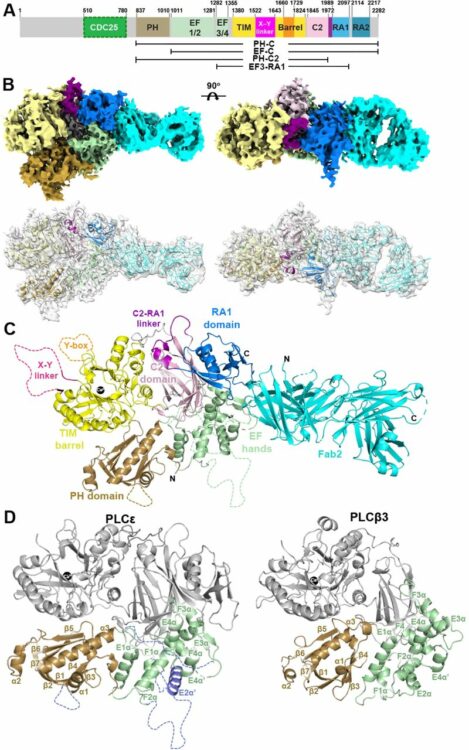
Cryo-EM Structure of Phospholipase Cε Defines N-terminal Domains and their Roles in Activity Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39314324,
title = {Cryo-EM Structure of Phospholipase Cε Defines N-terminal Domains and their Roles in Activity},
author = {Kadidia Samassekou and Elisabeth E Garland-Kuntz and Vaani Ohri and Isaac J Fisher and Satchal K Erramilli and Kaushik Muralidharan and Livia M Bogdan and Abigail M Gick and Anthony A Kossiakoff and Angeline M Lyon},
doi = {10.1101/2024.09.11.612521},
issn = {2692-8205},
year = {2024},
date = {2024-09-01},
urldate = {2024-09-01},
journal = {bioRxiv},
abstract = {Phospholipase Cε (PLCε) increases intracellular Ca and protein kinase C (PKC) activity in the cardiovascular system in response to stimulation of G protein coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs). The ability of PLCε to respond to these diverse inputs is due, in part, to multiple, conformationally dynamic regulatory domains. However, this heterogeneity has also limited structural studies of the lipase to either individual domains or its catalytic core. Here, we report the 3.9 Å reconstruction of the largest fragment of PLCε to date in complex with an antigen binding fragment (Fab). The structure reveals that PLCε contains a pleckstrin homology (PH) domain and four tandem EF hands, including subfamily-specific insertions and intramolecular interactions with the catalytic core. The structure, together with a model of the holoenzyme, suggest that part of the N-terminus and PH domain form a continuous surface that could engage cytoplasmic leaflets of the plasma and perinuclear membranes, contributing to activity. Functional characterization of this surface confirm it is critical for maximum basal and G protein-stimulated activities. This study provides new insights into the autoinhibited, basal conformation of PLCε and the first mechanistic insights into how it engages cellular membranes for activity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}