Publications
View by year
- Publications: 2025
- Publications: 2024
- Publications: 2023
- Publications: 2022
- Publications: 2021
- Publications: 2020
- Publications: 2019
- Publications: 2018
- Publications: 2017
- Publications: 2016
- Publications: 2015
- Publications: 2014
- Publications: 2013
- Publications: 2012
Search
Search for author, keywords, or any other term.
Mann, Samuel I; Lin, Zhi; Tan, Sophia K; Zhu, Jiaqi; Widel, Zachary X W; Bakanas, Ian; Mansergh, Jarrett P; Liu, Rui; Kelly, Mark J S; Wu, Yibing; Wells, James A; Therien, Michael J; DeGrado, William F
De Novo Design of Proteins That Bind Naphthalenediimides, Powerful Photooxidants with Tunable Photophysical Properties Journal Article
In: J Am Chem Soc, 2025, ISSN: 1520-5126.
@article{pmid39982408,
title = {De Novo Design of Proteins That Bind Naphthalenediimides, Powerful Photooxidants with Tunable Photophysical Properties},
author = {Samuel I Mann and Zhi Lin and Sophia K Tan and Jiaqi Zhu and Zachary X W Widel and Ian Bakanas and Jarrett P Mansergh and Rui Liu and Mark J S Kelly and Yibing Wu and James A Wells and Michael J Therien and William F DeGrado},
doi = {10.1021/jacs.4c18151},
issn = {1520-5126},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {J Am Chem Soc},
abstract = { protein design provides a framework to test our understanding of protein function and build proteins with cofactors and functions not found in nature. Here, we report the design of proteins designed to bind powerful photooxidants and the evaluation of the use of these proteins to generate diffusible small-molecule reactive species. Because excited-state dynamics are influenced by the dynamics and hydration of a photooxidant's environment, it was important to not only design a binding site but also to evaluate its dynamic properties. Thus, we used computational design in conjunction with molecular dynamics (MD) simulations to design a protein, designated NBP (DI inding rotein), that held a naphthalenediimide (NDI), a powerful photooxidant, in a programmable molecular environment. Solution NMR confirmed the structure of the complex. We evaluated two NDI cofactors in this protein using ultrafast pump-probe spectroscopy to evaluate light-triggered intra- and intermolecular electron transfer function. Moreover, we demonstrated the utility of this platform to activate multiple molecular probes for protein labeling.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

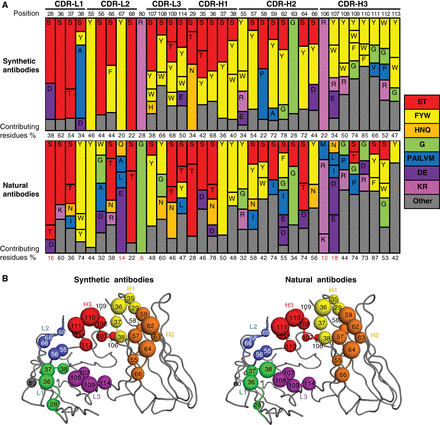
Gorelik, Maryna; Miersch, Shane; Sidhu, Sachdev S
Structural Survey of Antigen Recognition by Synthetic Human Antibodies Journal Article
In: Cold Spring Harb Protoc, vol. 2025, no. 2, pp. pdb.over107759, 2025, ISSN: 1559-6095.
@article{pmid38594044,
title = {Structural Survey of Antigen Recognition by Synthetic Human Antibodies},
author = {Maryna Gorelik and Shane Miersch and Sachdev S Sidhu},
doi = {10.1101/pdb.over107759},
issn = {1559-6095},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {Cold Spring Harb Protoc},
volume = {2025},
number = {2},
pages = {pdb.over107759},
abstract = {Synthetic antibody libraries have been used extensively to isolate and optimize antibodies. To generate these libraries, the immunological diversity and the antibody framework(s) that supports it outside of the binding regions are carefully designed/chosen to ensure favorable functional and biophysical properties. In particular, minimalist, single-framework synthetic libraries pioneered by our group have yielded a vast trove of antibodies to a broad array of antigens. Here, we review their systematic and iterative development to provide insights into the design principles that make them a powerful tool for drug discovery. In addition, the ongoing accumulation of crystal structures of antigen-binding fragment (Fab)-antigen complexes generated with synthetic antibodies enables a deepening understanding of the structural determinants of antigen recognition and usage of immunoglobulin sequence diversity, which can assist in developing new strategies for antibody and library optimization. Toward this, we also survey here the structural landscape of a comprehensive and unbiased set of 50 distinct complexes derived from these libraries and compare it to a similar set of natural antibodies with the goal of better understanding how each achieves molecular recognition and whether opportunities exist for iterative improvement of synthetic libraries. From this survey, we conclude that despite the minimalist strategies used for design of these synthetic antibody libraries, the overall structural interaction landscapes are highly similar to natural repertoires. We also found, however, some key differences that can help guide the iterative design of new synthetic libraries via the introduction of positionally tailored diversity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
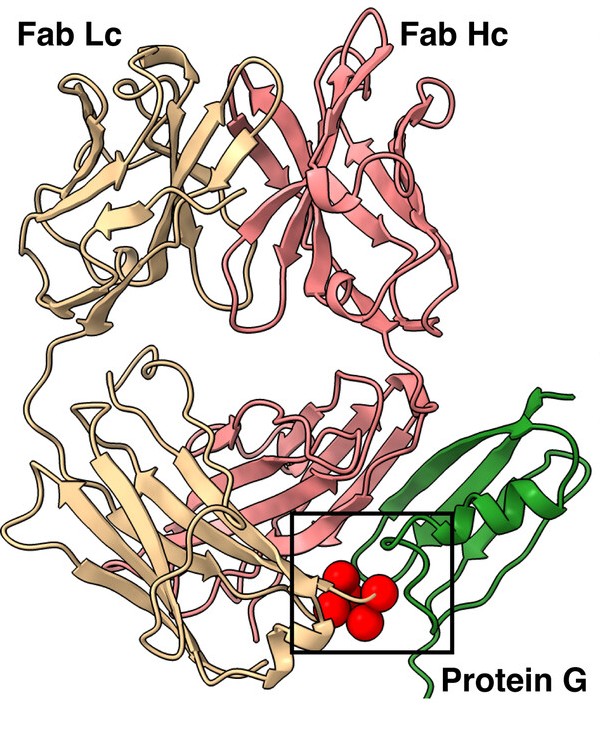
Slezak, Tomasz; O'Leary, Kelly M; Li, Jinyang; Rohaim, Ahmed; Davydova, Elena K; Kossiakoff, Anthony A
Engineered protein G variants for multifunctional antibody-based assemblies Journal Article
In: Protein Sci, vol. 34, no. 2, pp. e70019, 2025, ISSN: 1469-896X.
@article{pmid39865354,
title = {Engineered protein G variants for multifunctional antibody-based assemblies},
author = {Tomasz Slezak and Kelly M O'Leary and Jinyang Li and Ahmed Rohaim and Elena K Davydova and Anthony A Kossiakoff},
doi = {10.1002/pro.70019},
issn = {1469-896X},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {Protein Sci},
volume = {34},
number = {2},
pages = {e70019},
abstract = {We have developed a portfolio of antibody-based modules that can be prefabricated as standalone units and snapped together in plug-and-play fashion to create uniquely powerful multifunctional assemblies. The basic building blocks are derived from multiple pairs of native and modified Fab scaffolds and protein G (PG) variants engineered by phage display to introduce high pair-wise specificity. The variety of possible Fab-PG pairings provides a highly orthogonal system that can be exploited to perform challenging cell biology operations in a straightforward manner. The simplest manifestation allows multiplexed antigen detection using PG variants fused to fluorescently labeled SNAP-tags. Moreover, Fabs can be readily attached to a PG-Fc dimer module which acts as the core unit to produce plug-and-play IgG-like assemblies, and the utility can be further expanded to produce bispecific analogs using the "knobs into holes" strategy. These core PG-Fc dimer modules can be made and stored in bulk to produce off-the-shelf customized IgG entities in minutes, not days or weeks by just adding a Fab with the desired antigen specificity. In another application, the bispecific modalities form the building block for fabricating potent bispecific T-cell engagers (BiTEs), demonstrating their efficacy in cancer cell-killing assays. Additionally, the system can be adapted to include commercial antibodies as building blocks, greatly increasing the target space. Crystal structure analysis reveals that a few strategically positioned interactions engender the specificity between the Fab-PG variant pairs, requiring minimal changes to match the scaffolds for different possible combinations. This plug-and-play platform offers a user-friendly and versatile approach to enhance the functionality of antibody-based reagents in cell biology research.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
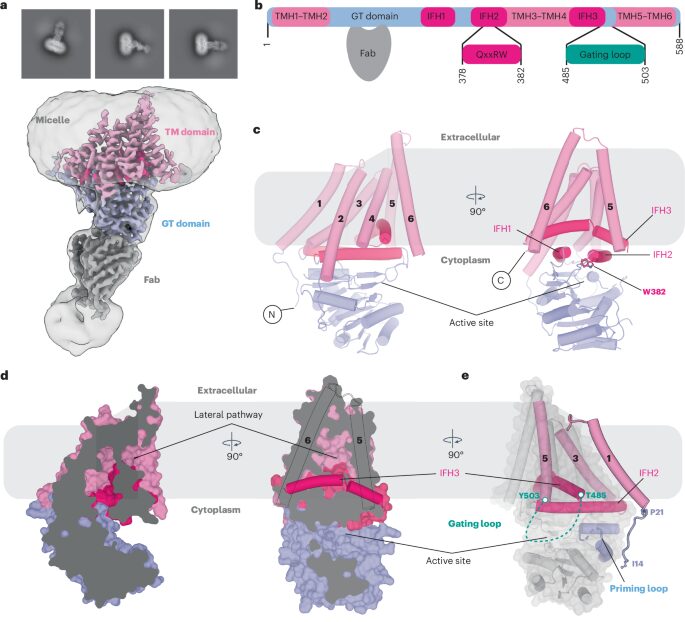
Górniak, Ireneusz; Stephens, Zachery; Erramilli, Satchal K; Gawda, Tomasz; Kossiakoff, Anthony A; Zimmer, Jochen
Structural insights into translocation and tailored synthesis of hyaluronan Journal Article
In: Nat Struct Mol Biol, vol. 32, no. 1, pp. 161–171, 2025, ISSN: 1545-9985.
@article{pmid39322765,
title = {Structural insights into translocation and tailored synthesis of hyaluronan},
author = {Ireneusz Górniak and Zachery Stephens and Satchal K Erramilli and Tomasz Gawda and Anthony A Kossiakoff and Jochen Zimmer},
doi = {10.1038/s41594-024-01389-1},
issn = {1545-9985},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Nat Struct Mol Biol},
volume = {32},
number = {1},
pages = {161--171},
abstract = {Hyaluronan (HA) is an essential component of the vertebrate extracellular matrix. It is a heteropolysaccharide of N-acetylglucosamine (GlcNAc) and glucuronic acid (GlcA) reaching several megadaltons in healthy tissues. HA is synthesized and translocated in a coupled reaction by HA synthase (HAS). Here, structural snapshots of HAS provide insights into HA biosynthesis, from substrate recognition to HA elongation and translocation. We monitor the extension of a GlcNAc primer with GlcA, reveal the coordination of the uridine diphosphate product by a conserved gating loop and capture the opening of a translocation channel to coordinate a translocating HA polymer. Furthermore, we identify channel-lining residues that modulate HA product lengths. Integrating structural and biochemical analyses suggests an avenue for polysaccharide engineering based on finely tuned enzymatic activity and HA coordination.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Rabe, Daniel C; Choudhury, Adarsh; Lee, Dasol; Luciani, Evelyn G; Ho, Uyen K; Clark, Alex E; Glasgow, Jeffrey E; Veiga, Sara; Michaud, William A; Capen, Diane; Flynn, Elizabeth A; Hartmann, Nicola; Garretson, Aaron F; Muzikansky, Alona; Goldberg, Marcia B; Kwon, Douglas S; Yu, Xu; Carlin, Aaron F; Theriault, Yves; Wells, James A; Lennerz, Jochen K; Lai, Peggy S; Rabi, Sayed Ali; Hoang, Anh N; Boland, Genevieve M; Stott, Shannon L
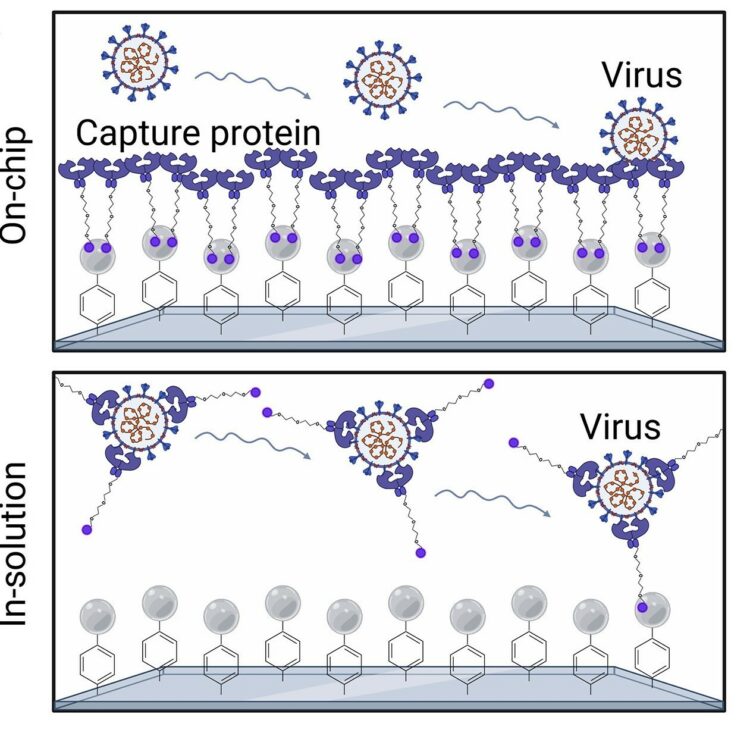
Ultrasensitive detection of intact SARS-CoV-2 particles in complex biofluids using microfluidic affinity capture Journal Article
In: Sci Adv, vol. 11, no. 2, pp. eadh1167, 2025, ISSN: 2375-2548.
@article{pmid39792670,
title = {Ultrasensitive detection of intact SARS-CoV-2 particles in complex biofluids using microfluidic affinity capture},
author = {Daniel C Rabe and Adarsh Choudhury and Dasol Lee and Evelyn G Luciani and Uyen K Ho and Alex E Clark and Jeffrey E Glasgow and Sara Veiga and William A Michaud and Diane Capen and Elizabeth A Flynn and Nicola Hartmann and Aaron F Garretson and Alona Muzikansky and Marcia B Goldberg and Douglas S Kwon and Xu Yu and Aaron F Carlin and Yves Theriault and James A Wells and Jochen K Lennerz and Peggy S Lai and Sayed Ali Rabi and Anh N Hoang and Genevieve M Boland and Shannon L Stott},
doi = {10.1126/sciadv.adh1167},
issn = {2375-2548},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Sci Adv},
volume = {11},
number = {2},
pages = {eadh1167},
abstract = {Measuring virus in biofluids is complicated by confounding biomolecules coisolated with viral nucleic acids. To address this, we developed an affinity-based microfluidic device for specific capture of intact severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Our approach used an engineered angiotensin-converting enzyme 2 to capture intact virus from plasma and other complex biofluids. Our device leverages a staggered herringbone pattern, nanoparticle surface coating, and processing conditions to achieve detection of as few as 3 viral copies per milliliter. We further validated our microfluidic assay on 103 plasma, 36 saliva, and 29 stool samples collected from unique patients with COVID-19, showing SARS-CoV-2 detection in 72% of plasma samples. Longitudinal monitoring in the plasma revealed our device's capacity for ultrasensitive detection of active viral infections over time. Our technology can be adapted to target other viruses using relevant cell entry molecules for affinity capture. This versatility underscores the potential for widespread application in viral load monitoring and disease management.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Ngo, Wayne; Peukes, Julia; Baldwin, Alisha; Xue, Zhiwei Wayne; Hwang, Sidney; Stickels, Robert R; Lin, Zhi; Satpathy, Ansuman T; Wells, James A; Schekman, Randy; Nogales, Eva; Doudna, Jennifer A
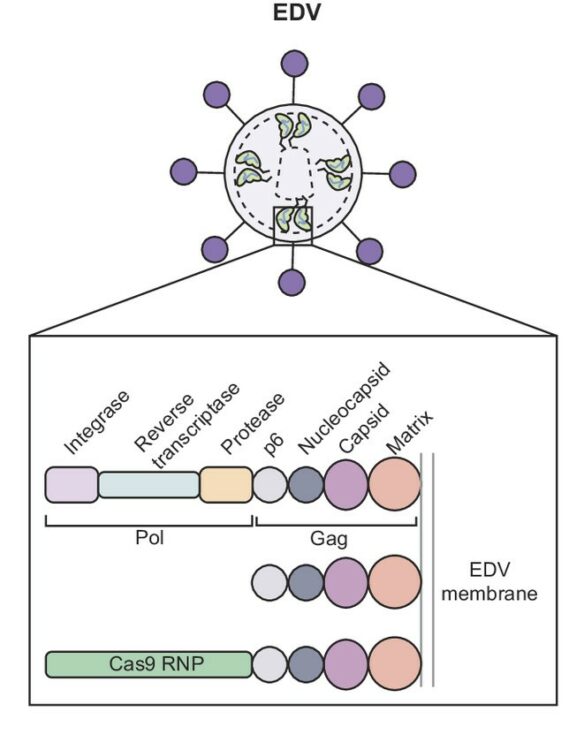
Mechanism-guided engineering of a minimal biological particle for genome editing Journal Article
In: Proc Natl Acad Sci U S A, vol. 122, no. 1, pp. e2413519121, 2025, ISSN: 1091-6490.
@article{pmid39793042,
title = {Mechanism-guided engineering of a minimal biological particle for genome editing},
author = {Wayne Ngo and Julia Peukes and Alisha Baldwin and Zhiwei Wayne Xue and Sidney Hwang and Robert R Stickels and Zhi Lin and Ansuman T Satpathy and James A Wells and Randy Schekman and Eva Nogales and Jennifer A Doudna},
doi = {10.1073/pnas.2413519121},
issn = {1091-6490},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Proc Natl Acad Sci U S A},
volume = {122},
number = {1},
pages = {e2413519121},
abstract = {The widespread application of genome editing to treat and cure disease requires the delivery of genome editors into the nucleus of target cells. Enveloped delivery vehicles (EDVs) are engineered virally derived particles capable of packaging and delivering CRISPR-Cas9 ribonucleoproteins (RNPs). However, the presence of lentiviral genome encapsulation and replication proteins in EDVs has obscured the underlying delivery mechanism and precluded particle optimization. Here, we show that Cas9 RNP nuclear delivery is independent of the native lentiviral capsid structure. Instead, EDV-mediated genome editing activity corresponds directly to the number of nuclear localization sequences on the Cas9 enzyme. EDV structural analysis using cryo-electron tomography and small molecule inhibitors guided the removal of ~80% of viral residues, creating a minimal EDV (miniEDV) that retains full RNP delivery capability. MiniEDVs are 25% smaller yet package equivalent amounts of Cas9 RNPs relative to the original EDVs and demonstrated increased editing in cell lines and therapeutically relevant primary human T cells. These results show that virally derived particles can be streamlined to create efficacious genome editing delivery vehicles with simpler production and manufacturing.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Kordon, Szymon P; Cechova, Kristina; Bandekar, Sumit J; Leon, Katherine; Dutka, Przemysław; Siffer, Gracie; Kossiakoff, Anthony A; Vafabakhsh, Reza; Araç, Demet
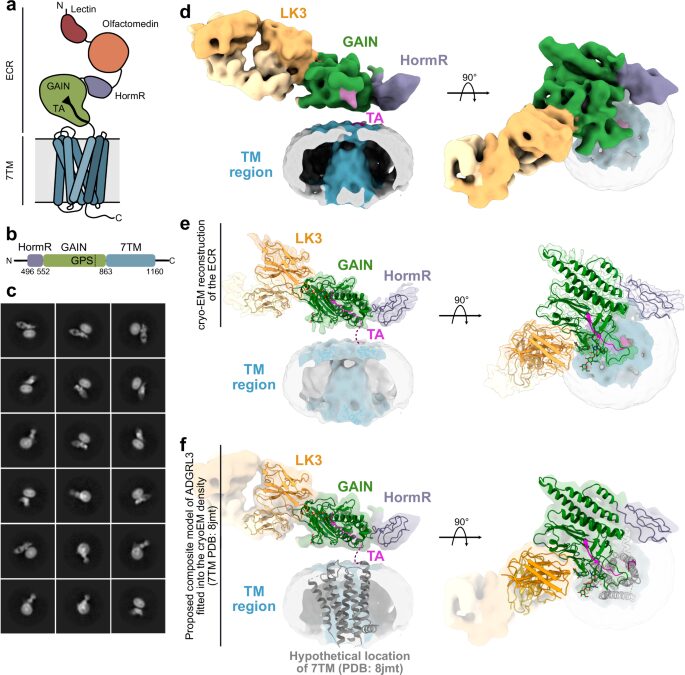
Conformational coupling between extracellular and transmembrane domains modulates holo-adhesion GPCR function Journal Article
In: Nat Commun, vol. 15, no. 1, pp. 10545, 2024, ISSN: 2041-1723.
@article{pmid39627215,
title = {Conformational coupling between extracellular and transmembrane domains modulates holo-adhesion GPCR function},
author = {Szymon P Kordon and Kristina Cechova and Sumit J Bandekar and Katherine Leon and Przemysław Dutka and Gracie Siffer and Anthony A Kossiakoff and Reza Vafabakhsh and Demet Araç},
doi = {10.1038/s41467-024-54836-4},
issn = {2041-1723},
year = {2024},
date = {2024-12-01},
urldate = {2024-12-01},
journal = {Nat Commun},
volume = {15},
number = {1},
pages = {10545},
abstract = {Adhesion G Protein-Coupled Receptors (aGPCRs) are key cell-adhesion molecules involved in numerous physiological functions. aGPCRs have large multi-domain extracellular regions (ECRs) containing a conserved GAIN domain that precedes their seven-pass transmembrane domain (7TM). Ligand binding and mechanical force applied on the ECR regulate receptor function. However, how the ECR communicates with the 7TM remains elusive, because the relative orientation and dynamics of the ECR and 7TM within a holoreceptor is unclear. Here, we describe the cryo-EM reconstruction of an aGPCR, Latrophilin3/ADGRL3, and reveal that the GAIN domain adopts a parallel orientation to the transmembrane region and has constrained movement. Single-molecule FRET experiments unveil three slow-exchanging FRET states of the ECR relative to the transmembrane region within the holoreceptor. GAIN-targeted antibodies, and cancer-associated mutations at the GAIN-7TM interface, alter FRET states, cryo-EM conformations, and receptor signaling. Altogether, this data demonstrates conformational and functional coupling between the ECR and 7TM, suggesting an ECR-mediated mechanism for aGPCR activation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Martyn, Gregory D; Kalagiri, Rajasree; Veggiani, Gianluca; Stanfield, Robyn L; Choudhuri, Indrani; Sala, Margaux; Meisenhelder, Jill; Chen, Chao; Biswas, Avik; Levy, Ronald M; Lyumkis, Dmitry; Wilson, Ian A; Hunter, Tony; Sidhu, Sachdev S
Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39574610,
title = {Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody},
author = {Gregory D Martyn and Rajasree Kalagiri and Gianluca Veggiani and Robyn L Stanfield and Indrani Choudhuri and Margaux Sala and Jill Meisenhelder and Chao Chen and Avik Biswas and Ronald M Levy and Dmitry Lyumkis and Ian A Wilson and Tony Hunter and Sachdev S Sidhu},
doi = {10.1101/2024.11.04.621849},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {Histidine phosphorylation (pHis) is a non-canonical post-translational modification (PTM) that is historically understudied due to a lack of robust reagents that are required for its investigation, such as high affinity pHis-specific antibodies. Engineering pHis-specific antibodies is very challenging due to the labile nature of the phosphoramidate (P-N) bond and the stringent requirements for selective recognition of the two isoforms, 1-phosphohistidine (1-pHis) and 3-phosphohistidine (3-pHis). Here, we present a strategy for engineering of antibodies for detection of native 3-pHis targets. Specifically, we humanized the rabbit SC44-8 anti-3-pTza (a stable 3-pHis mimetic) mAb into a scaffold (herein referred to as hSC44) that was suitable for phage display. We then constructed six unique Fab phage-displayed libraries using the hSC44 scaffold and selected high affinity 3-pHis binders. Our selection strategy was carefully designed to enrich antibodies that bound 3-pHis with high affinity and had specificity for 3-pHis versus 3-pTza. hSC44.20N32F, the best engineered antibody, has an ~10-fold higher affinity for 3-pHis than the parental hSC44. Eleven new Fab structures, including the first reported antibody-pHis peptide structures were solved by X-ray crystallography. Structural and quantum mechanical calculations provided molecular insights into 3-pHis and 3-pTza discrimination by different hSC44 variants and their affinity increase obtained through engineering. Furthermore, we demonstrate the utility of these newly developed high-affinity 3-pHis-specific antibodies for recognition of pHis proteins in mammalian cells by immunoblotting and immunofluorescence staining. Overall, our work describes a general method for engineering PTM-specific antibodies and provides a set of novel antibodies for further investigations of the role of 3-pHis in cell biology.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

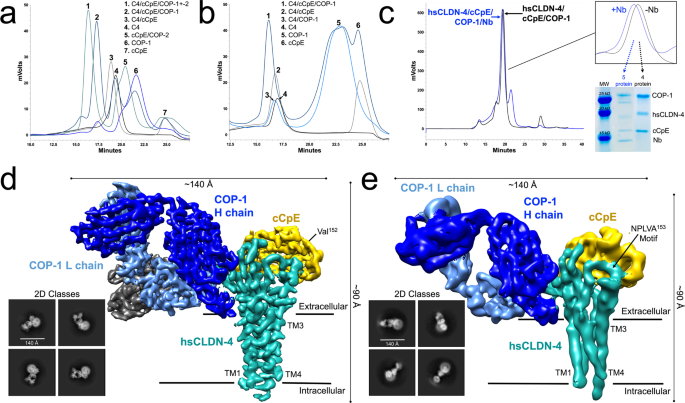
Rathnayake, Sewwandi S; Erramilli, Satchal K; Kossiakoff, Anthony A; Vecchio, Alex J
Cryo-EM structures of Clostridium perfringens enterotoxin bound to its human receptor, claudin-4 Journal Article
In: Structure, vol. 32, no. 11, pp. 1936–1951.e5, 2024, ISSN: 1878-4186.
@article{pmid39383874,
title = {Cryo-EM structures of Clostridium perfringens enterotoxin bound to its human receptor, claudin-4},
author = {Sewwandi S Rathnayake and Satchal K Erramilli and Anthony A Kossiakoff and Alex J Vecchio},
doi = {10.1016/j.str.2024.09.015},
issn = {1878-4186},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {Structure},
volume = {32},
number = {11},
pages = {1936--1951.e5},
abstract = {Clostridium perfringens enterotoxin (CpE) causes prevalent and deadly gastrointestinal disorders. CpE binds to receptors called claudins on the apical surfaces of small intestinal epithelium. Claudins normally regulate paracellular transport but are hijacked from doing so by CpE and are instead led to form claudin/CpE complexes. Claudin/CpE complexes are the building blocks of oligomeric β-barrel pores that penetrate the plasma membrane and induce gut cytotoxicity. Here, we present the structures of CpE in complex with its native claudin receptor in humans, claudin-4, using cryogenic electron microscopy. The structures reveal the architecture of the claudin/CpE complex, the residues used in binding, the orientation of CpE relative to the membrane, and CpE-induced changes to claudin-4. Further, structures and modeling allude to the biophysical procession from claudin/CpE complexes to cytotoxic β-barrel pores during pathogenesis. In full, this work proposes a model of claudin/CpE assembly and provides strategies to obstruct its formation to treat CpE diseases.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Ogbu, Chinemerem P; Mandriota, Alexandria M; Liu, Xiangdong; de Las Alas, Mason; Kapoor, Srajan; Choudhury, Jagrity; Kossiakoff, Anthony A; Duffey, Michael E; Vecchio, Alex J
Biophysical Basis of Paracellular Barrier Modulation by a Pan-Claudin-Binding Molecule Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39605593,
title = {Biophysical Basis of Paracellular Barrier Modulation by a Pan-Claudin-Binding Molecule},
author = {Chinemerem P Ogbu and Alexandria M Mandriota and Xiangdong Liu and Mason de Las Alas and Srajan Kapoor and Jagrity Choudhury and Anthony A Kossiakoff and Michael E Duffey and Alex J Vecchio},
doi = {10.1101/2024.11.10.622873},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {Claudins are a 27-member protein family that form and fortify specialized cell contacts in endothelium and epithelium called tight junctions. Tight junctions restrict paracellular transport across tissues by forming molecular barriers between cells. Claudin-binding molecules thus hold promise for modulating tight junction permeability to deliver drugs or as therapeutics to treat tight junction-linked disease. The development of claudin-binding molecules, however, is hindered by their intractability and small targetable surfaces. Here, we determine that a synthetic antibody fragment (sFab) we developed binds directly to 10 claudin subtypes with nanomolar affinity by targeting claudin's paracellular-exposed surface. Application of this sFab to cells that model intestinal epithelium show that it opens the paracellular barrier comparable to a known, but application limited, tight junction modulator. This novel pan-claudin-binding molecule can probe claudin or tight junction structure and holds potential as a broad modulator of tight junction permeability for basic or translational applications.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
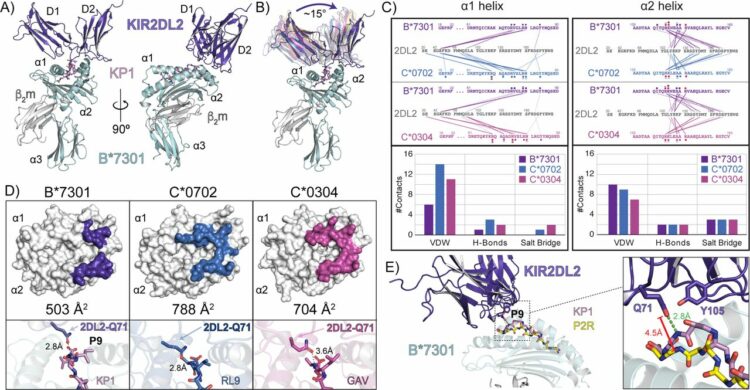
Ross, Philipp; Hilton, Hugo G; Lodwick, Jane; Slezak, Tomasz; Guethlein, Lisbeth A; McMurtrey, Curtis P; Han, Alex S; Nielsen, Morten; Yong, Daniel; Dulberger, Charles L; Nolan, Kristof T; Roy, Sobhan; Castro, Caitlin D; Hildebrand, William H; Zhao, Minglei; Kossiakoff, Anthony; Parham, Peter; Adams, Erin J
Molecular characterization of the archaic HLA-B*73:01 allele reveals presentation of a unique peptidome and skewed engagement by KIR2DL2 Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39651149,
title = {Molecular characterization of the archaic HLA-B*73:01 allele reveals presentation of a unique peptidome and skewed engagement by KIR2DL2},
author = {Philipp Ross and Hugo G Hilton and Jane Lodwick and Tomasz Slezak and Lisbeth A Guethlein and Curtis P McMurtrey and Alex S Han and Morten Nielsen and Daniel Yong and Charles L Dulberger and Kristof T Nolan and Sobhan Roy and Caitlin D Castro and William H Hildebrand and Minglei Zhao and Anthony Kossiakoff and Peter Parham and Erin J Adams},
doi = {10.1101/2024.11.25.625330},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {HLA class I alleles of archaic origin may have been retained in modern humans because they provide immunity against diseases to which archaic humans had evolved resistance. According to this model, archaic introgressed alleles were somehow distinct from those that evolved in African populations. Here we show that HLA-B*73:01, a rare allotype with putative archaic origins, has a relatively rare peptide binding motif with an unusually long-tailed peptide length distribution. We also find that HLA-B*73:01 combines a restricted and unique peptidome with high-cell surface expression, characteristics that make it well-suited to combat one or a number of closely-related pathogens. Furthermore, a crystal structure of HLA-B*73:01 in complex with KIR2DL2 highlights differences from previously solved structures with HLA-C molecules. These molecular characteristics distinguish HLA-B*73:01 from other HLA class I alleles previously investigated and may have provided early modern human migrants that inherited this allele with a selective advantage as they colonized Europe and Asia.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
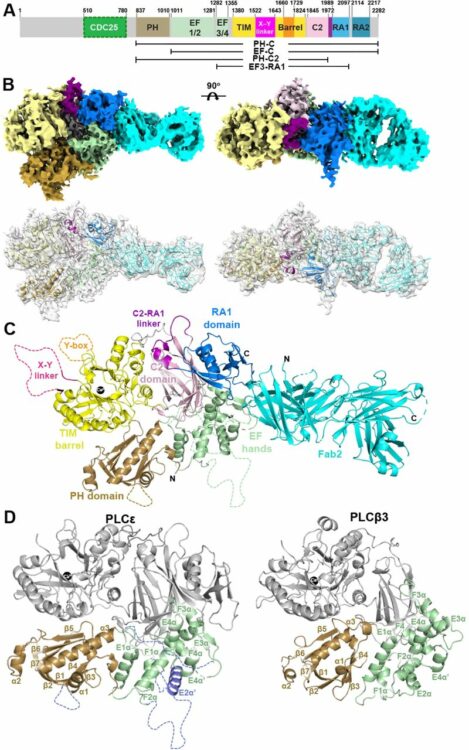
Samassekou, Kadidia; Garland-Kuntz, Elisabeth E; Ohri, Vaani; Fisher, Isaac J; Erramilli, Satchal K; Muralidharan, Kaushik; Bogdan, Livia M; Gick, Abigail M; Kossiakoff, Anthony A; Lyon, Angeline M
Cryo-EM Structure of Phospholipase Cε Defines N-terminal Domains and their Roles in Activity Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39314324,
title = {Cryo-EM Structure of Phospholipase Cε Defines N-terminal Domains and their Roles in Activity},
author = {Kadidia Samassekou and Elisabeth E Garland-Kuntz and Vaani Ohri and Isaac J Fisher and Satchal K Erramilli and Kaushik Muralidharan and Livia M Bogdan and Abigail M Gick and Anthony A Kossiakoff and Angeline M Lyon},
doi = {10.1101/2024.09.11.612521},
issn = {2692-8205},
year = {2024},
date = {2024-09-01},
urldate = {2024-09-01},
journal = {bioRxiv},
abstract = {Phospholipase Cε (PLCε) increases intracellular Ca and protein kinase C (PKC) activity in the cardiovascular system in response to stimulation of G protein coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs). The ability of PLCε to respond to these diverse inputs is due, in part, to multiple, conformationally dynamic regulatory domains. However, this heterogeneity has also limited structural studies of the lipase to either individual domains or its catalytic core. Here, we report the 3.9 Å reconstruction of the largest fragment of PLCε to date in complex with an antigen binding fragment (Fab). The structure reveals that PLCε contains a pleckstrin homology (PH) domain and four tandem EF hands, including subfamily-specific insertions and intramolecular interactions with the catalytic core. The structure, together with a model of the holoenzyme, suggest that part of the N-terminus and PH domain form a continuous surface that could engage cytoplasmic leaflets of the plasma and perinuclear membranes, contributing to activity. Functional characterization of this surface confirm it is critical for maximum basal and G protein-stimulated activities. This study provides new insights into the autoinhibited, basal conformation of PLCε and the first mechanistic insights into how it engages cellular membranes for activity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
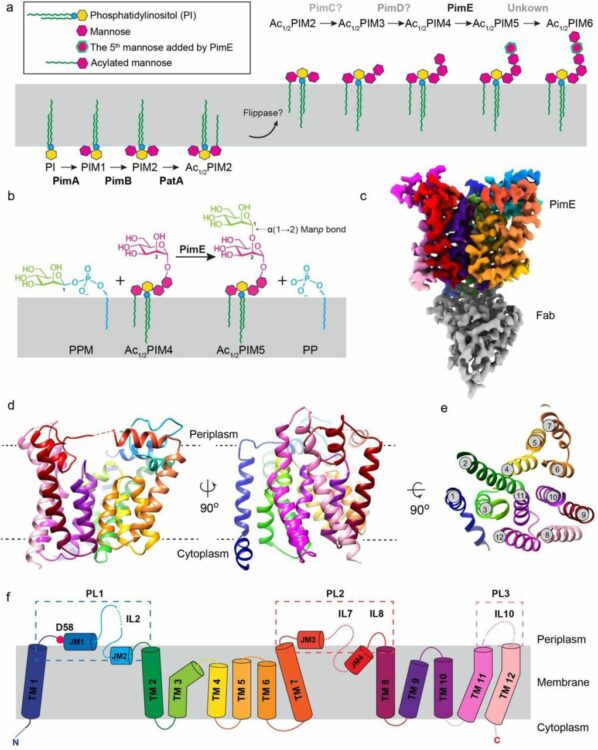
Liu, Yaqi; Brown, Chelsea M; Borges, Nuno; Nobre, Rodrigo N; Erramilli, Satchal; Dufrisne, Meagan Belcher; Kloss, Brian; Giacometti, Sabrina; Esteves, Ana M; Timoteo, Cristina G; Tokarz, Piotr; Cater, Rosemary; Morita, Yasu S; Kossiakoff, Anthony A; Santos, Helena; Stansfeld, Phillip J; Nygaard, Rie; Mancia, Filippo
Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39345506,
title = {Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE},
author = {Yaqi Liu and Chelsea M Brown and Nuno Borges and Rodrigo N Nobre and Satchal Erramilli and Meagan Belcher Dufrisne and Brian Kloss and Sabrina Giacometti and Ana M Esteves and Cristina G Timoteo and Piotr Tokarz and Rosemary Cater and Yasu S Morita and Anthony A Kossiakoff and Helena Santos and Phillip J Stansfeld and Rie Nygaard and Filippo Mancia},
doi = {10.1101/2024.09.17.613550},
issn = {2692-8205},
year = {2024},
date = {2024-09-01},
urldate = {2024-09-01},
journal = {bioRxiv},
abstract = {Tuberculosis (TB), exceeded in mortality only by COVID-19 among global infectious diseases, is caused by Mycobacterium tuberculosis (Mtb). The pathogenicity of Mtb is largely attributed to its complex cell envelope, which includes a class of glycolipids called phosphatidyl-myo-inositol mannosides (PIMs), found uniquely in mycobacteria and its related corynebacterineae. These glycolipids maintain the integrity of the mycobacterial cell envelope, regulate its permeability, and mediate host-pathogen interactions. PIMs consist of a phosphatidyl-myo-inositol core decorated with one to six mannose residues and up to four acyl chains. The mannosyltransferase PimE catalyzes the transfer of the fifth PIM mannose residue from a polyprenyl phosphate-mannose (PPM) donor. This step in the biosynthesis of higher-order PIMs contributes to the proper assembly and function of the mycobacterial cell envelope; however, the structural basis for substrate recognition and the catalytic mechanism of PimE remain poorly understood. Here, we present the cryo-electron microscopy (cryo-EM) structures of PimE from Mycobacterium abscessus captured in its apo form and in a product-bound complex with the reaction product Ac1PIM5 and the by-product polyprenyl phosphate (PP), determined at 3.0 Å and 3.5 Å, respectively. The structures reveal the active site within a distinctive binding cavity that accommodates both donor and acceptor substrates/products. Within the cavity, we identified residues involved in substrate coordination and catalysis, which we confirmed through in vitro enzymatic assays and further validated by in vivo complementation experiments. Molecular dynamics simulations were applied to identify the access pathways and the dynamics involved in substrate binding. Integrating structural, biochemical, genetic, and computational experiments, our study provides comprehensive insights into how PimE functions, opening potential avenues for development of novel anti-TB therapeutics.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
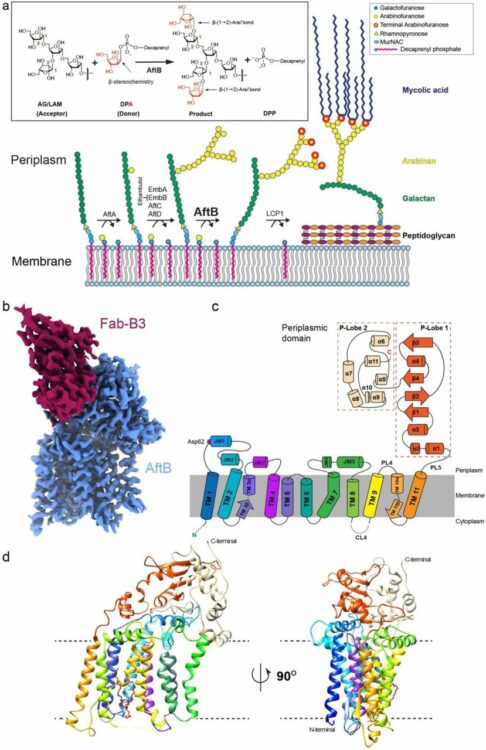
Liu, Yaqi; Brown, Chelsea M; Erramilli, Satchal; Su, Yi-Chia; Tseng, Po-Sen; Wang, Yu-Jen; Duong, Nam Ha; Tokarz, Piotr; Kloss, Brian; Han, Cheng-Ruei; Chen, Hung-Yu; Rodrigues, Jose; Archer, Margarida; Lowary, Todd L; Kossiakoff, Anthony A; Stansfeld, Phillip J; Nygaard, Rie; Mancia, Filippo
Structural insights into terminal arabinosylation biosynthesis of the mycobacterial cell wall arabinan Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39345558,
title = {Structural insights into terminal arabinosylation biosynthesis of the mycobacterial cell wall arabinan},
author = {Yaqi Liu and Chelsea M Brown and Satchal Erramilli and Yi-Chia Su and Po-Sen Tseng and Yu-Jen Wang and Nam Ha Duong and Piotr Tokarz and Brian Kloss and Cheng-Ruei Han and Hung-Yu Chen and Jose Rodrigues and Margarida Archer and Todd L Lowary and Anthony A Kossiakoff and Phillip J Stansfeld and Rie Nygaard and Filippo Mancia},
doi = {10.1101/2024.09.17.613533},
issn = {2692-8205},
year = {2024},
date = {2024-09-01},
urldate = {2024-09-01},
journal = {bioRxiv},
abstract = {The emergence of drug-resistant strains exacerbates the global challenge of tuberculosis caused by Mycobacterium tuberculosis (Mtb). Central to the pathogenicity of Mtb is its complex cell envelope, which serves as a barrier against both immune system and pharmacological attacks. Two key components of this envelope, arabinogalactan (AG) and lipoarabinomannan (LAM) are complex polysaccharides that contain integral arabinan domains important for cell wall structural and functional integrity. The arabinofuranosyltransferase AftB terminates the synthesis of these arabinan domains by catalyzing the addition of the addition of β-(1→2)-linked terminal arabinofuranose residues. Here, we present the cryo-EM structures of Mycobacterium chubuense AftB in its apo and donor substrate analog-bound form, determined to 2.9 Å and 3.4 Å resolution, respectively. Our structures reveal that AftB has a GT-C fold transmembrane (TM) domain comprised of eleven TM helices and a periplasmic cap domain. AftB has an irregular tube-shaped cavity that bridges the two proposed substrate binding sites. By integrating structural analysis, biochemical assays, and molecular dynamics simulations, we elucidate the molecular basis of the reaction mechanism of AftB and propose a model for catalysis.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
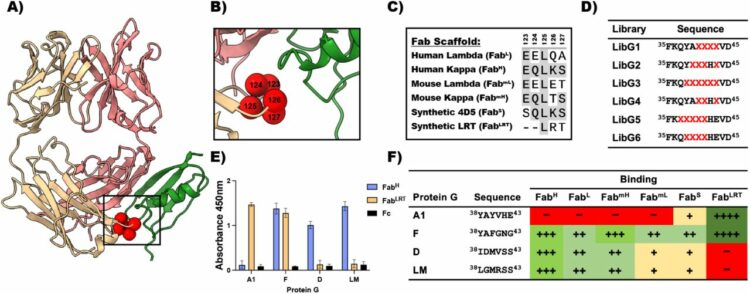
Slezak, Tomasz; O'Leary, Kelly M; Li, Jinyang; Rohaim, Ahmed; Davydova, Elena K; Kossiakoff, Anthony A
Engineered Protein-G variants for plug-and-play applications Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39211271,
title = {Engineered Protein-G variants for plug-and-play applications},
author = {Tomasz Slezak and Kelly M O'Leary and Jinyang Li and Ahmed Rohaim and Elena K Davydova and Anthony A Kossiakoff},
doi = {10.1101/2024.08.06.606809},
issn = {2692-8205},
year = {2024},
date = {2024-08-01},
urldate = {2024-08-01},
journal = {bioRxiv},
abstract = {We have developed a portfolio of antibody-based modules that can be prefabricated as standalone units and snapped together in plug-and-play fashion to create uniquely powerful multifunctional assemblies. The basic building blocks are derived from multiple pairs of native and modified Fab scaffolds and protein G (PG) variants engineered by phage display to introduce high pair-wise specificity. The variety of possible Fab-PG pairings provides a highly orthogonal system that can be exploited to perform challenging cell biology operations in a straightforward manner. The simplest manifestation allows multiplexed antigen detection using PG variants fused to fluorescently labeled SNAP-tags. Moreover, Fabs can be readily attached to a PG-Fc dimer module which acts as the core unit to produce plug-and-play IgG-like assemblies, and the utility can be further expanded to produce bispecific analogs using the "knobs into holes" strategy. These core PG-Fc dimer modules can be made and stored in bulk to produce off-the-shelf customized IgG entities in minutes, not days or weeks by just adding a Fab with the desired antigen specificity. In another application, the bispecific modalities form the building block for fabricating potent Bispecific T-cell Engagers (BiTEs), demonstrating their efficacy in cancer cell-killing assays. Additionally, the system can be adapted to include commercial antibodies as building blocks, greatly increasing the target space. Crystal structure analysis reveals that a few strategically positioned interactions engender the specificity between the Fab-PG variant pairs, requiring minimal changes to match the scaffolds for different possible combinations. This plug-and-play platform offers a user-friendly and versatile approach to enhance the functionality of antibody-based reagents in cell biology research.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
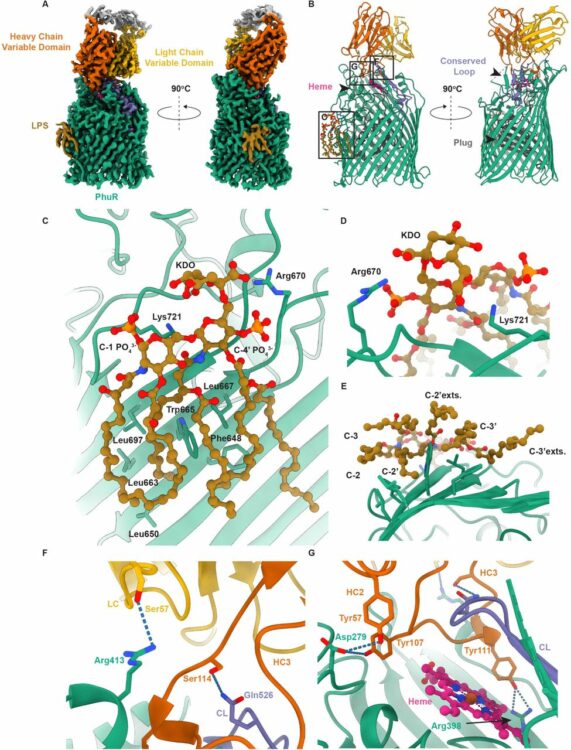
Knejski, Paweł P; Erramilli, Satchal K; Kossiakoff, Anthony A
Chaperone-assisted cryo-EM structure of PhuR reveals molecular basis for heme binding Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid37577460,
title = {Chaperone-assisted cryo-EM structure of PhuR reveals molecular basis for heme binding},
author = {Paweł P Knejski and Satchal K Erramilli and Anthony A Kossiakoff},
doi = {10.1101/2023.08.01.551527},
issn = {2692-8205},
year = {2024},
date = {2024-08-01},
urldate = {2024-08-01},
journal = {bioRxiv},
abstract = {Pathogenic bacteria, such as , depend on scavenging heme for the acquisition of iron, an essential nutrient. The TonB-dependent transporter (TBDT) PhuR is the major heme uptake protein in clinical isolates. However, a comprehensive understanding of heme recognition and TBDT transport mechanisms, especially PhuR, remains limited. In this study, we employed single-particle cryogenic electron microscopy (cryo-EM) and a phage display-generated synthetic antibody (sAB) as a fiducial marker to enable the determination of a high-resolution (2.5 Å) structure of PhuR with a bound heme. Notably, the structure reveals iron coordination by Y529 on a conserved extracellular loop, shedding light on the role of tyrosine in heme binding. Biochemical assays and negative-stain EM demonstrated that the sAB specifically targets the heme-bound state of PhuR. These findings provide insights into PhuR's heme binding and offer a template for developing conformation-specific sABs against outer membrane proteins (OMPs) for structure-function investigations.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
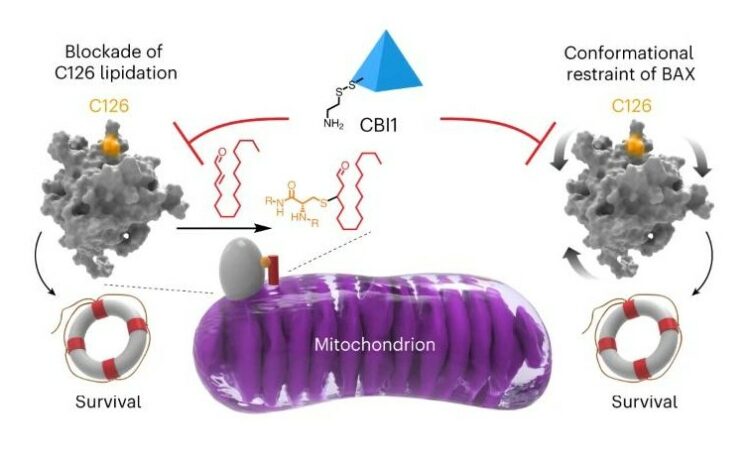
McHenry, Matthew W; Shi, Peiwen; Camara, Christina M; Cohen, Daniel T; Rettenmaier, T Justin; Adhikary, Utsarga; Gygi, Micah A; Yang, Ka; Gygi, Steven P; Wales, Thomas E; Engen, John R; Wells, James A; Walensky, Loren D
Covalent inhibition of pro-apoptotic BAX Journal Article
In: Nat Chem Biol, vol. 20, no. 8, pp. 1022–1032, 2024, ISSN: 1552-4469.
@article{pmid38233584,
title = {Covalent inhibition of pro-apoptotic BAX},
author = {Matthew W McHenry and Peiwen Shi and Christina M Camara and Daniel T Cohen and T Justin Rettenmaier and Utsarga Adhikary and Micah A Gygi and Ka Yang and Steven P Gygi and Thomas E Wales and John R Engen and James A Wells and Loren D Walensky},
doi = {10.1038/s41589-023-01537-6},
issn = {1552-4469},
year = {2024},
date = {2024-08-01},
urldate = {2024-08-01},
journal = {Nat Chem Biol},
volume = {20},
number = {8},
pages = {1022--1032},
abstract = {BCL-2-associated X protein (BAX) is a promising therapeutic target for activating or restraining apoptosis in diseases of pathologic cell survival or cell death, respectively. In response to cellular stress, BAX transforms from a quiescent cytosolic monomer into a toxic oligomer that permeabilizes the mitochondria, releasing key apoptogenic factors. The mitochondrial lipid trans-2-hexadecenal (t-2-hex) sensitizes BAX activation by covalent derivatization of cysteine 126 (C126). In this study, we performed a disulfide tethering screen to discover C126-reactive molecules that modulate BAX activity. We identified covalent BAX inhibitor 1 (CBI1) as a compound that selectively derivatizes BAX at C126 and inhibits BAX activation by triggering ligands or point mutagenesis. Biochemical and structural analyses revealed that CBI1 can inhibit BAX by a dual mechanism of action: conformational constraint and competitive blockade of lipidation. These data inform a pharmacologic strategy for suppressing apoptosis in diseases of unwanted cell death by covalent targeting of BAX C126.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
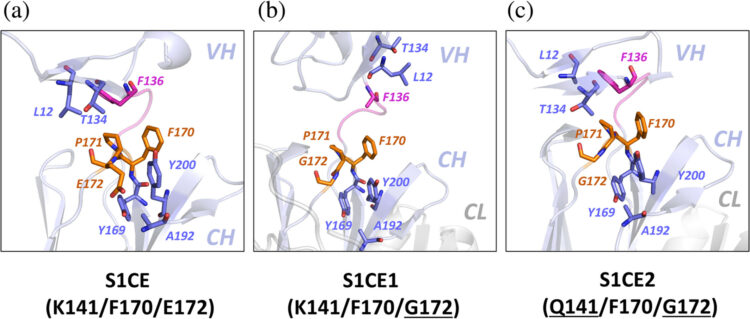
Bruce, Heather A.; Singer, Alexander U.; Blazer, Levi L.; Luu, Khanh; Ploder, Lynda; Pavlenco, Alevtina; Kurinov, Igor; Adams, Jarrett J.; Sidhu, Sachdev S.
In: Protein Science, vol. 33, no. 7, 2024, ISSN: 1469-896X.
@article{Bruce2024,
title = {Antigen‐binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones},
author = {Heather A. Bruce and Alexander U. Singer and Levi L. Blazer and Khanh Luu and Lynda Ploder and Alevtina Pavlenco and Igor Kurinov and Jarrett J. Adams and Sachdev S. Sidhu},
doi = {10.1002/pro.5081},
issn = {1469-896X},
year = {2024},
date = {2024-07-00},
urldate = {2024-07-00},
journal = {Protein Science},
volume = {33},
number = {7},
publisher = {Wiley},
abstract = {<jats:title>Abstract</jats:title><jats:p>It has been shown previously that a set of three modifications—termed S1, Crystal Kappa, and elbow—act synergistically to improve the crystallizability of an antigen‐binding fragment (Fab) framework. Here, we prepared a phage‐displayed library and performed crystallization screenings to identify additional substitutions—located near the heavy‐chain elbow region—which cooperate with the S1, Crystal Kappa, and elbow modifications to increase expression and improve crystallizability of the Fab framework even further. One substitution (K141Q) supports the signature Crystal Kappa‐mediated Fab:Fab crystal lattice packing interaction. Another substitution (E172G) improves the compatibility of the elbow modification with the Fab framework by alleviating some of the strain incurred by the shortened and bulkier elbow linker region. A third substitution (F170W) generates a split‐Fab conformation, resulting in a powerful crystal lattice packing interaction comprising the biological interaction interface between the variable heavy and light chain domains. In sum, we have used K141Q, E172G, and F170W substitutions—which complement the S1, Crystal Kappa, and elbow modifications—to generate a set of highly crystallizable Fab frameworks that can be used as chaperones to enable facile elucidation of Fab:antigen complex structures by x‐ray crystallography.</jats:p>},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
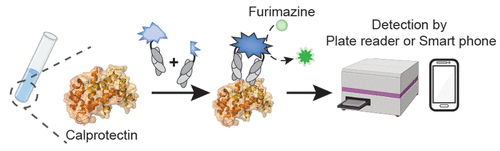
Lan, Tong; Slezak, Tomasz; Pu, Jinyue; Zinkus-Boltz, Julia; Adhikari, Sarbani; Pekow, Joel R; Taneja, Vibha; Zuniga, Joaquin; Gómez-García, Itzel A; Regino-Zamarripa, Nora; Ahmed, Mushtaq; Khader, Shabaana A; Rubin, David T; Kossiakoff, Anthony A; Dickinson, Bryan C
Development of Luminescent Biosensors for Calprotectin Journal Article
In: ACS Chem Biol, vol. 19, no. 6, pp. 1250–1259, 2024, ISSN: 1554-8937.
@article{pmid38843544,
title = {Development of Luminescent Biosensors for Calprotectin},
author = {Tong Lan and Tomasz Slezak and Jinyue Pu and Julia Zinkus-Boltz and Sarbani Adhikari and Joel R Pekow and Vibha Taneja and Joaquin Zuniga and Itzel A Gómez-García and Nora Regino-Zamarripa and Mushtaq Ahmed and Shabaana A Khader and David T Rubin and Anthony A Kossiakoff and Bryan C Dickinson},
doi = {10.1021/acschembio.4c00265},
issn = {1554-8937},
year = {2024},
date = {2024-06-01},
urldate = {2024-06-01},
journal = {ACS Chem Biol},
volume = {19},
number = {6},
pages = {1250--1259},
abstract = {Calprotectin, a metal ion-binding protein complex, plays a crucial role in the innate immune system and has gained prominence as a biomarker for various intestinal and systemic inflammatory and infectious diseases, including inflammatory bowel disease (IBD) and tuberculosis (TB). Current clinical testing methods rely on enzyme-linked immunosorbent assays (ELISAs), limiting accessibility and convenience. In this study, we introduce the Fab-Enabled Split-luciferase Calprotectin Assay (FESCA), a novel quantitative method for calprotectin measurement. FESCA utilizes two new fragment antigen binding proteins (Fabs), CP16 and CP17, that bind to different epitopes of the calprotectin complex. These Fabs are fused with split NanoLuc luciferase fragments, enabling the reconstitution of active luciferase upon binding to calprotectin either in solution or in varied immobilized assay formats. FESCA's output luminescence can be measured with standard laboratory equipment as well as consumer-grade cell phone cameras. FESCA can detect physiologically relevant calprotectin levels across various sample types, including serum, plasma, and whole blood. Notably, FESCA can detect abnormally elevated native calprotectin from TB patients. In summary, FESCA presents a convenient, low-cost, and quantitative method for assessing calprotectin levels in various biological samples, with the potential to improve the diagnosis and monitoring of inflammatory diseases, especially in at-home or point-of-care settings.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
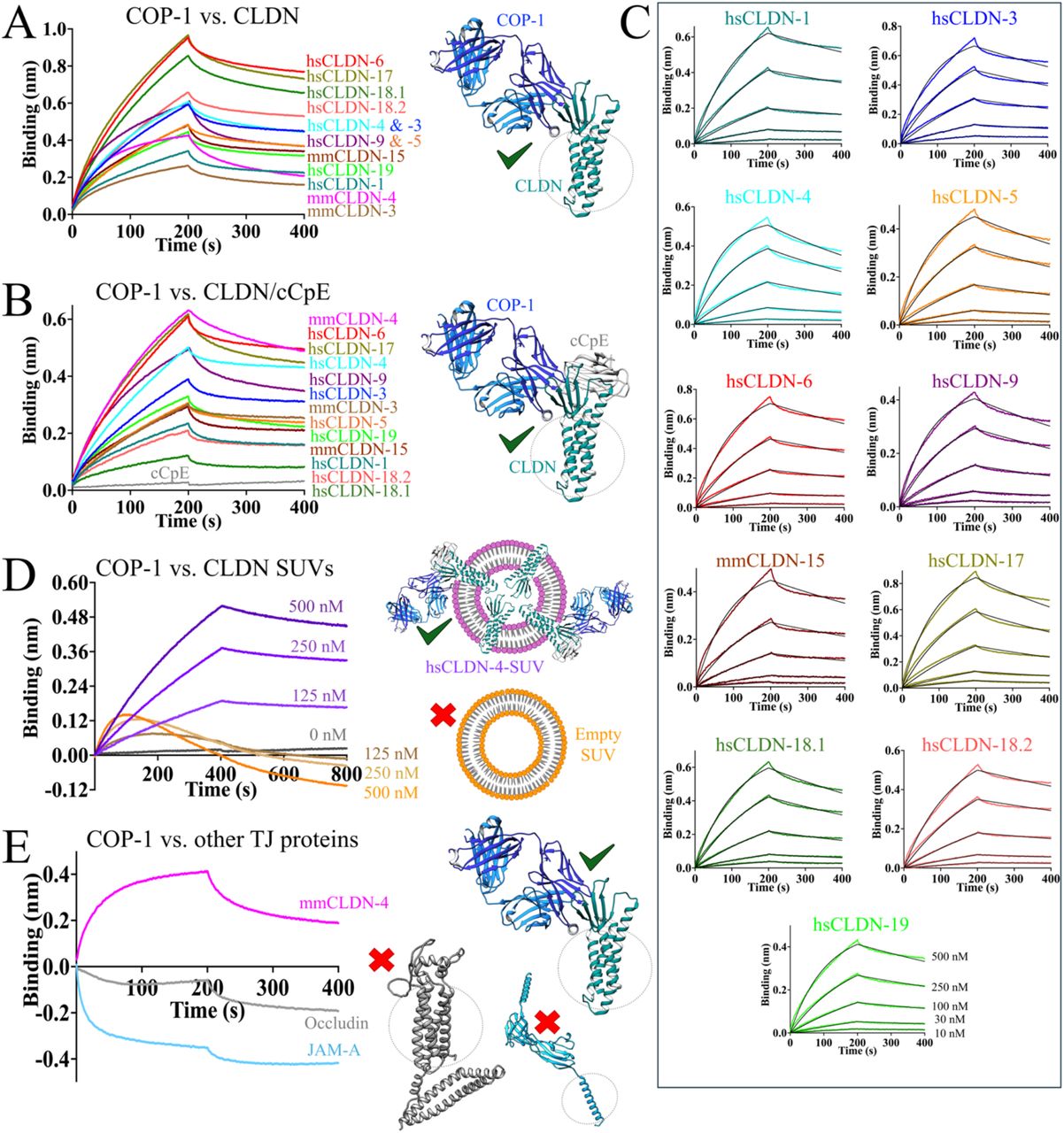
Erramilli, Satchal K; Dominik, Pawel K; Ogbu, Chinemerem P; Kossiakoff, Anthony A; Vecchio, Alex J
Structural and biophysical insights into targeting of claudin-4 by a synthetic antibody fragment Journal Article
In: Commun Biol, vol. 7, no. 1, pp. 733, 2024, ISSN: 2399-3642.
@article{pmid38886509,
title = {Structural and biophysical insights into targeting of claudin-4 by a synthetic antibody fragment},
author = {Satchal K Erramilli and Pawel K Dominik and Chinemerem P Ogbu and Anthony A Kossiakoff and Alex J Vecchio},
doi = {10.1038/s42003-024-06437-6},
issn = {2399-3642},
year = {2024},
date = {2024-06-01},
urldate = {2024-06-01},
journal = {Commun Biol},
volume = {7},
number = {1},
pages = {733},
abstract = {Claudins are a 27-member family of ~25 kDa membrane proteins that integrate into tight junctions to form molecular barriers at the paracellular spaces between endothelial and epithelial cells. As the backbone of tight junction structure and function, claudins are attractive targets for modulating tissue permeability to deliver drugs or treat disease. However, structures of claudins are limited due to their small sizes and physicochemical properties-these traits also make therapy development a challenge. Here we report the development of a synthetic antibody fragment (sFab) that binds human claudin-4 and the determination of a high-resolution structure of it bound to claudin-4/enterotoxin complexes using cryogenic electron microscopy. Structural and biophysical results reveal this sFabs mechanism of select binding to human claudin-4 over other homologous claudins and establish the ability of sFabs to bind hard-to-target claudins to probe tight junction structure and function. The findings provide a framework for tight junction modulation by sFabs for tissue-selective therapies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}