Publications: 2025
2025
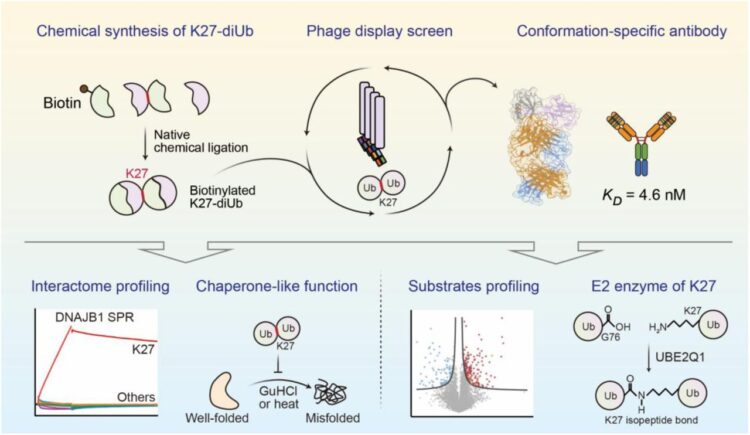
Han, Chengxiao; Weng, Yicheng; Zheng, Qingyun; Qu, Qian; Erramilli, Satchal K; Su, Zhen; Duan, Yujuan; Han, Yunxi; Zhai, Xiaoguo; Li, Jingxian; Kossiakoff, Anthony A; Pan, Man; Zhao, Minglei; Liu, Lei; Yu, Yuanyuan
Conformation-specific Antibody Deciphers K27-linked Ubiquitination in Chaperone-Mediated Proteostasis Journal Article
In: bioRxiv, 2025, ISSN: 2692-8205.
@article{pmid41446272,
title = {Conformation-specific Antibody Deciphers K27-linked Ubiquitination in Chaperone-Mediated Proteostasis},
author = {Chengxiao Han and Yicheng Weng and Qingyun Zheng and Qian Qu and Satchal K Erramilli and Zhen Su and Yujuan Duan and Yunxi Han and Xiaoguo Zhai and Jingxian Li and Anthony A Kossiakoff and Man Pan and Minglei Zhao and Lei Liu and Yuanyuan Yu},
doi = {10.64898/2025.12.18.695067},
issn = {2692-8205},
year = {2025},
date = {2025-12-01},
urldate = {2025-12-01},
journal = {bioRxiv},
abstract = {Lysine 27 (K27)-linked polyubiquitination plays critical yet incompletely defined roles in proteostasis, innate immunity, and disease progression; however, investigations into this process have long been hindered by its extremely low abundance and the lack of conformation-specific enrichment tools. Herein, we describe the development of a long-sought conformation-specific antibody, K27-IgG, which can selectively recognize-among all ubiquitin chain types-the unique architecture of K27-linked polyubiquitin (K27-polyUb) characterized by a distinct buried K27-isopeptide bond, with high affinity (KD = 4.66 nM). This antibody was derived from synthetic antibodies initially generated via phage display, using chemically synthesized K27-linked diubiquitin (K27-diUb) as the antigen. High-resolution co-crystal structures uncovered the unique K27-diUb interface targeted by these sAbs. Subsequent reformatting of these sAbs into a full-length human immunoglobulin G (IgG) scaffold yielded K27-IgG, notably exhibiting markedly enhanced affinity without compromising selectivity. Using K27-IgG as a tool, we achieved sensitive detection and immunoprecipitation (IP) of endogenous K27-polyUb in cells, and delineated the intracellular interaction landscape of K27-polyUb through complementary proteomic approaches. Two key findings emerged: 1) The molecular chaperone DNAJB1 is a specific reader of K27-linked ubiquitin chains (but not other linkages) and that K27-polyUb chains themselves exhibit chaperone-like activity, suggesting a novel mechanism by which K27-polyUb regulates chaperone-mediated proteostasis; 2) The E2 enzyme UBE2Q1 assembles K27-diUb, identifying it as a potential writer for this ubiquitin chain topology. Collectively, this study establishes K27-IgG as a robust tool for deciphering the K27-linked ubiquitin code, thereby opening new avenues for investigating the biological functions of K27-linked polyubiquitination.nnHIGHLIGHTS: First K27-linkage conformation-specific antibody with nanomolar affinity overcomes a major barrier in the field.K27-IgG unlocks functional mapping of the K27 ubiquitin landscape under proteotoxic stress.Molecular chaperone DNAJB1 is a selective "reader" of K27-linked ubiquitin chains.K27 chains possess intrinsic chaperone activity, enabling protein refolding and suppressing aggregation.E2 enzyme UBE2Q1 is a "writer" that directly assembles K27-linked ubiquitin chains.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
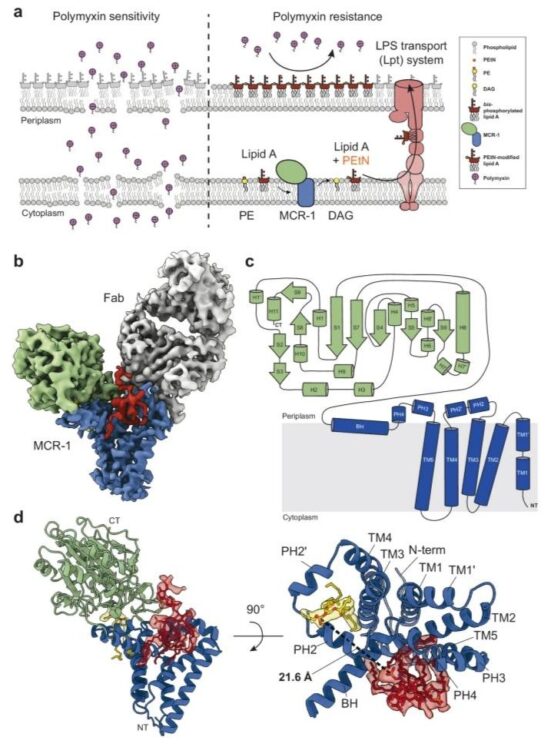
Zinkle, Allen P; Batista, Mariana Bunoro; Herrera, Carmen M; Erramilli, Satchal K; Kloss, Brian; Ashraf, Khuram U; Nosol, Kamil; Zhang, Guozhi; Cater, Rosemary J; Marty, Michael T; Kossiakoff, Anthony A; Trent, M Stephen; Nygaard, Rie; Stansfeld, Phillip J; Mancia, Filippo
Mechanistic basis of antimicrobial resistance mediated by the phosphoethanolamine transferase MCR-1 Journal Article
In: Nat Commun, vol. 16, no. 1, pp. 10516, 2025, ISSN: 2041-1723.
@article{pmid41298376,
title = {Mechanistic basis of antimicrobial resistance mediated by the phosphoethanolamine transferase MCR-1},
author = {Allen P Zinkle and Mariana Bunoro Batista and Carmen M Herrera and Satchal K Erramilli and Brian Kloss and Khuram U Ashraf and Kamil Nosol and Guozhi Zhang and Rosemary J Cater and Michael T Marty and Anthony A Kossiakoff and M Stephen Trent and Rie Nygaard and Phillip J Stansfeld and Filippo Mancia},
doi = {10.1038/s41467-025-65515-3},
issn = {2041-1723},
year = {2025},
date = {2025-11-01},
urldate = {2025-11-01},
journal = {Nat Commun},
volume = {16},
number = {1},
pages = {10516},
abstract = {Polymyxins are used to treat infections caused by multidrug-resistant Gram-negative bacteria. They are cationic peptides that target the negatively charged lipid A component of lipopolysaccharides, disrupting the outer membrane and lysing the cell. Polymyxin resistance is conferred by inner-membrane enzymes, such as phosphoethanolamine transferases, which add positively charged phosphoethanolamine to lipid A. Here, we present the structure of MCR-1, a plasmid-encoded phosphoethanolamine transferase, in its liganded form. The phosphatidylethanolamine donor substrate is bound near the active site in the periplasmic domain, and lipid A is bound over 20 Å away, within the transmembrane region. Integrating structural, biochemical, and drug-resistance data with computational analyses, we propose a two-state model in which the periplasmic domain rotates to bring the active site to lipid A, near the preferential phosphate modification site for MCR-1. This enzymatic mechanism may be generally applicable to other phosphoform transferases with large, globular soluble domains.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
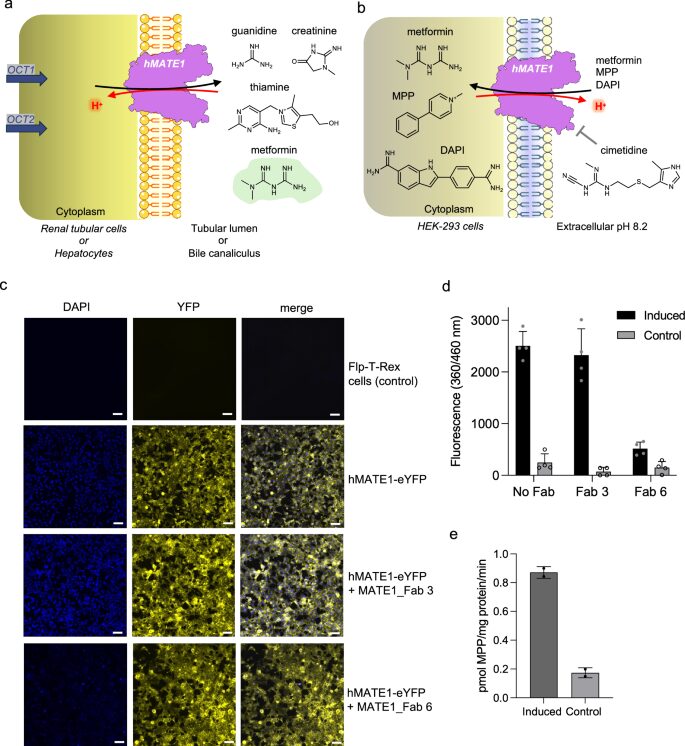
Romane, Ksenija; Peteani, Giulia; Mukherjee, Somnath; Kowal, Julia; Rossi, Lorenzo; Hou, Jingkai; Kossiakoff, Anthony A; Lemmin, Thomas; Locher, Kaspar P
Structural basis of drug recognition by human MATE1 transporter Journal Article
In: Nat Commun, vol. 16, no. 1, pp. 9444, 2025, ISSN: 2041-1723.
@article{pmid41145429,
title = {Structural basis of drug recognition by human MATE1 transporter},
author = {Ksenija Romane and Giulia Peteani and Somnath Mukherjee and Julia Kowal and Lorenzo Rossi and Jingkai Hou and Anthony A Kossiakoff and Thomas Lemmin and Kaspar P Locher},
doi = {10.1038/s41467-025-64490-z},
issn = {2041-1723},
year = {2025},
date = {2025-10-28},
urldate = {2025-10-01},
journal = {Nat Commun},
volume = {16},
number = {1},
pages = {9444},
abstract = {Human MATE1 (multidrug and toxin extrusion protein 1) is highly expressed in the kidney and liver, where it mediates the final step in the excretion of a broad range of cationic drugs, including the antidiabetic drug metformin, into the urine and bile. This transport process is essential for drug clearance and also affects therapeutic efficacy. To understand the molecular basis of drug recognition by hMATE1, we determined cryo-electron microscopy structures of the transporter in complex with the substrates 1-methyl-4-phenylpyridinium (MPP) and metformin and with the inhibitor cimetidine. The structures reveal a shared binding site located in a negatively charged pocket in the C-lobe of the protein. We functionally validated key interactions using radioactivity-based cellular uptake assays using hMATE1 mutants. Molecular dynamics simulations provide insights into the different binding modes and dynamic behaviour of the ligands within the pocket. Collectively, these findings define the structural basis of hMATE1 substrate specificity and shed light on its role in drug transport and drug-drug interactions.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Zhao, Fangzhu; Wu, Yan; Schaefer, Kaitlin; Zhang, Yun; Miao, Kun; Yao, Zi; Ganjave, Snehal D; Kumru, Kaan; Peters-Clarke, Trenton M; Inague, Alex; Olzmann, James A; Leung, Kevin K; Wells, James A
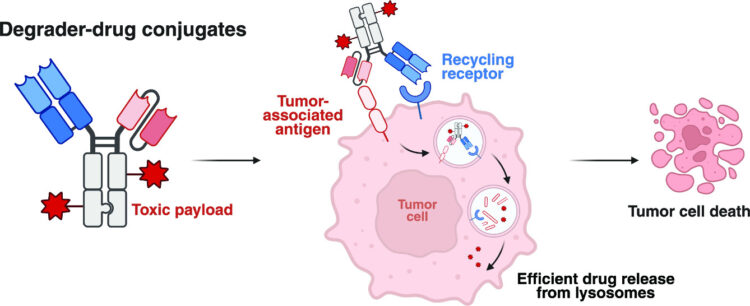
Hijacking Extracellular Targeted Protein Degrader-Drug Conjugates for Enhanced Drug Delivery Journal Article
In: J Am Chem Soc, 2025, ISSN: 1520-5126.
@article{pmid41091621,
title = {Hijacking Extracellular Targeted Protein Degrader-Drug Conjugates for Enhanced Drug Delivery},
author = {Fangzhu Zhao and Yan Wu and Kaitlin Schaefer and Yun Zhang and Kun Miao and Zi Yao and Snehal D Ganjave and Kaan Kumru and Trenton M Peters-Clarke and Alex Inague and James A Olzmann and Kevin K Leung and James A Wells},
doi = {10.1021/jacs.5c15047},
issn = {1520-5126},
year = {2025},
date = {2025-10-15},
urldate = {2025-10-01},
journal = {J Am Chem Soc},
abstract = {Antibody-based therapeutics encompass diverse modalities for targeting tumor cells. Among these, antibody-drug conjugates (ADCs) and extracellular targeted protein degradation (eTPD) specifically depend on efficient lysosomal trafficking for activity. A major limitation of ADCs is their reliance on antigens with efficient internalization, while eTPD approaches, although capable of trafficking diverse targets to lysosomes, lack cytotoxic potency. To address this, we developed degrader-drug conjugates (DDCs), leveraging the endocytic and recycling activities of eTPD to enhance lysosomal delivery. We utilized fast internalizers, the low-density lipoprotein receptor (LDLR) and the chemokine receptor (CXCR7), to enhance lysosomal delivery. LDLR-based degraders enabled efficient and selective degradation of diverse extracellular membrane proteins, while DDCs with cytotoxic payload enhanced cytotoxicity compared to conventional ADCs . This dual modality addresses key challenges of inadequate internalization in conventional ADCs and cytotoxic potency in current eTPD strategies. Our findings demonstrate that DDCs provide additional optionality for developing next-generation antibody therapeutics with broader utility and improved efficacy in cancer treatment.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Mohona, Surovi; Shakya, Anil K; Singh, Suruchi; Kearns, Fiona L; Jemison, Kezia; Erramilli, Satchal; Dey, Debajit; Qing, Enya; Jennings, Benjamin C; Doray, Balraj; Kossiakoff, Anthony A; Amaro, Rommie E; Klose, Thomas; Gallagher, Tom; Hasan, S Saif
An unconventional HxD motif orchestrates coatomer-dependent coronavirus morphogenesis Journal Article
In: bioRxiv, 2025, ISSN: 2692-8205.
@article{pmid41279991,
title = {An unconventional HxD motif orchestrates coatomer-dependent coronavirus morphogenesis},
author = {Surovi Mohona and Anil K Shakya and Suruchi Singh and Fiona L Kearns and Kezia Jemison and Satchal Erramilli and Debajit Dey and Enya Qing and Benjamin C Jennings and Balraj Doray and Anthony A Kossiakoff and Rommie E Amaro and Thomas Klose and Tom Gallagher and S Saif Hasan},
doi = {10.1101/2025.10.16.682669},
issn = {2692-8205},
year = {2025},
date = {2025-10-01},
urldate = {2025-10-01},
journal = {bioRxiv},
abstract = {Assembly of infectious coronaviruses requires spike (S) protein trafficking by host coatomer, typically via a dibasic signal in the S cytoplasmic tail. However, the human embecoviruses HKU1 and OC43, as well as the model virus MHV, lack this motif. Here we identify a conserved His-x-Asp (HxD) sequence that functions as an unconventional coatomer-binding signal. Structural and biochemical analyses show that the MHV HxD motif engages coatomer subunits through distinct conformations, while cellular imaging demonstrates its role in directing S to assembly sites with the viral M-protein. Disruption of HxD-coatomer interactions impairs S incorporation and provokes compensatory viral adaptations, including emergence of a canonical dibasic motif or mutations in M-protein. Electron microscopy further reveals profound alterations in virion surface architecture. These findings uncover HxD as a previously unrecognized coatomer-targeting motif, highlighting an unexpected flexibility in coronavirus assembly pathways and broadening understanding of the cellular machinery that shapes coronavirus morphogenesis.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Adams, Jarrett J.; Blazer, Levi L.; Chung, Jacky; Karimi, Minoo; Davidson, Taylor; Bruce, Heather A.; Singer, Alexander U.; Yang, Ning; Cardarelli, Lia; Pot, Isabelle; Colombo, Luigi; Huang, Lily Jun‐shen; Ma, Yue; Michnick, Stephen W.; Moe, Orson W.; Sidhu, Sachdev S.
An asymmetric tetrabody is a potent and efficacious agonist of the erythropoietin receptor in vitro and in vivo Journal Article
In: Protein Science, vol. 34, no. 10, 2025, ISSN: 1469-896X.
@article{Adams2025,
title = {An asymmetric tetrabody is a potent and efficacious agonist of the erythropoietin receptor in vitro and in vivo},
author = {Jarrett J. Adams and Levi L. Blazer and Jacky Chung and Minoo Karimi and Taylor Davidson and Heather A. Bruce and Alexander U. Singer and Ning Yang and Lia Cardarelli and Isabelle Pot and Luigi Colombo and Lily Jun‐shen Huang and Yue Ma and Stephen W. Michnick and Orson W. Moe and Sachdev S. Sidhu},
doi = {10.1002/pro.70292},
issn = {1469-896X},
year = {2025},
date = {2025-09-17},
urldate = {2025-10-00},
journal = {Protein Science},
volume = {34},
number = {10},
publisher = {Wiley},
abstract = {Abstract: Erythropoietin (EPO) initiates EPO receptor (EPOR) signaling in hematopoietic cells by binding to an asymmetric EPOR dimer through two different sites. We engineered dimeric diabody‐Fc (Db‐Fc) fusion proteins that appeared to act as potent agonists of human EPOR in cell proliferation assays. However, detailed analysis of their oligomeric forms revealed that the predominant Db‐Fc species bound EPOR with high affinity but failed to induce cell proliferation. Instead, a minor oligomeric form, identified as a putative tetrabody (Tb) fused to two Fc domains (Tb‐Fc), proved to be the minimal active form. The existence of a tetrameric agonist was further supported by crystallography, which revealed an asymmetric Tb structure. Additionally, the structure of an antigen‐binding fragment (Fab) bound to EPOR revealed an epitope distinct from the EPO binding sites, and structural modeling showed that engagement of two of the four binding sites on the Tb could form an asymmetric EPOR dimer nearly identical to the active conformation recruited by EPO. In a knock‐in mouse model, where mouse EPOR was replaced by human EPOR, purified Tb‐Fc stimulated erythropoiesis with greater potency, efficacy, and duration than darbepoetin, a recombinant EPO that is the leading therapeutic erythropoiesis‐stimulating agent (ESA). Collectively, these findings demonstrate that asymmetric tetravalent antibodies such as Tb‐Fc represent promising next‐generation ESAs that provide enhanced potency, efficacy, and durability. Moreover, they may reduce the oncogenic and cardiovascular risks associated with the pleiotropy of EPO.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Wadhwa, Anju; Johnson, Haley; Bobba, Kondapa Naidu; Bidkar, Anil P; Mayne, Ellis; Rampersaud, Sham; Mandal, Kamal; Barpanda, Abhilash; Prudhvi, Sanjana; Kang, Amrik S; Greenland, Nancy; Balitzer, Dana; Peter, Robin; Raveendran, Athira; Naik, Shubhankar; Basak, Megha; Kasap, Corynn; Huebner, Juwita; Alvarez, Marina Lopez; Lee, Sanghee; Steri, Veronica; Adams, Jarret J; Sidhu, Sachdev S; Wilson, David M; Seo, Youngho; VanBrocklin, Henry F; Logan, Aaron C; Wiita, Arun P; Flavell, Robert R
Effective imaging and treatment of Acute Myeloid Leukemia with radiotheranostics targeting the activated conformation of integrin-βeta2 Journal Article
In: bioRxiv, 2025, ISSN: 2692-8205.
@article{pmid40909732,
title = {Effective imaging and treatment of Acute Myeloid Leukemia with radiotheranostics targeting the activated conformation of integrin-βeta2},
author = {Anju Wadhwa and Haley Johnson and Kondapa Naidu Bobba and Anil P Bidkar and Ellis Mayne and Sham Rampersaud and Kamal Mandal and Abhilash Barpanda and Sanjana Prudhvi and Amrik S Kang and Nancy Greenland and Dana Balitzer and Robin Peter and Athira Raveendran and Shubhankar Naik and Megha Basak and Corynn Kasap and Juwita Huebner and Marina Lopez Alvarez and Sanghee Lee and Veronica Steri and Jarret J Adams and Sachdev S Sidhu and David M Wilson and Youngho Seo and Henry F VanBrocklin and Aaron C Logan and Arun P Wiita and Robert R Flavell},
doi = {10.1101/2025.08.21.671206},
issn = {2692-8205},
year = {2025},
date = {2025-08-01},
urldate = {2025-08-01},
journal = {bioRxiv},
abstract = {There remains an unmet clinical need for improved treatment strategies in Acute Myeloid Leukemia (AML). Although radiopharmaceutical therapies targeting non-cancer-selective antigens have shown promise in AML, their clinical utility is often limited by prolonged bone marrow suppression. Using a unique proteomics-based strategy, we recently identified the active conformation of integrin-β2 (aITGB2) as a novel, tumor-selective target for AML. Importantly, this conformational epitope is expressed widely on AML cells but minimally on normal marrow progenitors/healthy tissues. Here we first confirmed widespread aITGB2 expression on AML tumors that was largely independent of tumor genotype or prior therapeutic regimen. We developed diagnostic and therapeutic radiopharmaceuticals targeting aITGB2 utilizing a conformation-specific antibody (clone 7065). PET/CT imaging with Zr and Ce-labeled 7065 in AML models revealed high target-mediated uptake, greater than that compared to standard of care [ F]-FDG. PET/CT imaging with [ Zr]DFO*-7065 showed reduced binding to normal bone marrow and immune cells in humanized immune system mice compared to [ Zr]DFO*-anti-CD33. For therapy, we developed [ Ac]Macropa-PEG -7065 using an optimized chelator-linker combination. Treatment with [ Ac]Macropa-PEG -7065 in Nomo-1 and PDX AML disseminated models delayed tumor growth and improved overall survival compared to controls, including [ Ac]DOTA-anti-CD33, a clinical stage-radioimmunotherapy under evaluation in AML. Relapsed tumors demonstrated persistent aITGB2 expression, supporting continued development of fractionated dosing schemes, and proteomics analysis indicated activation of TCA cycle and carbon metabolism pathways, consistent with therapy-induced stress responses. These findings highlight [ Zr]DFO*-7065 and [ Ac]Macropa-7065 as a promising aITGB2-targeted theranostic pair with potential for imaging and treatment in future clinical translation.nnONE SENTENCE SUMMARY: This study demonstrates promising preclinical efficacy of aITGB2-targeted radiotheranostics for selective imaging and therapy in AML.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
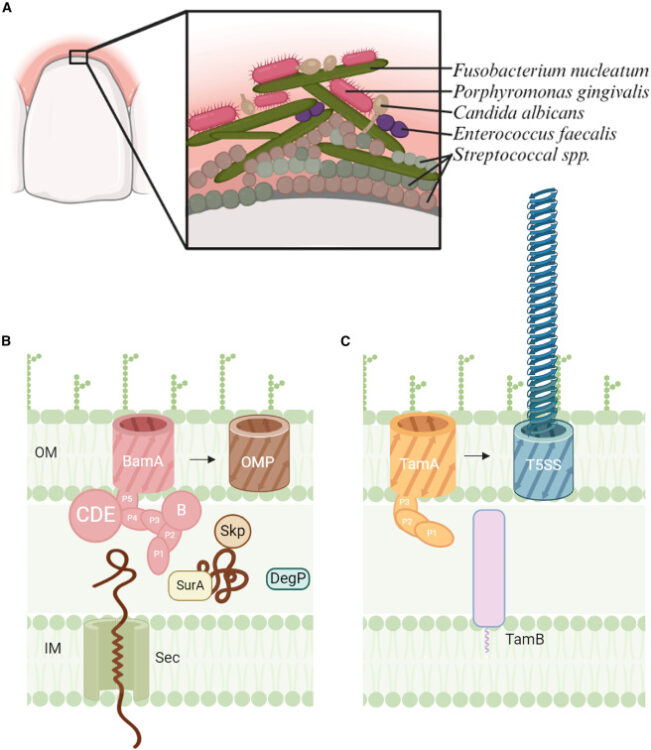
Cottom, Claire Overly; Heinz, Eva; Erramilli, Satchal; Kossiakoff, Anthony; Slade, Daniel J; Noinaj, Nicholas
Characterization of the OMP biogenesis machinery in Fusobacterium nucleatum Journal Article
In: Structure, 2025, ISSN: 1878-4186.
@article{pmid40897170,
title = {Characterization of the OMP biogenesis machinery in Fusobacterium nucleatum},
author = {Claire Overly Cottom and Eva Heinz and Satchal Erramilli and Anthony Kossiakoff and Daniel J Slade and Nicholas Noinaj},
doi = {10.1016/j.str.2025.08.008},
issn = {1878-4186},
year = {2025},
date = {2025-08-01},
urldate = {2025-08-01},
journal = {Structure},
abstract = {F. nucleatum is a Gram-negative bacteria that causes oral infections and is linked to colorectal cancer. Pathogenicity relies on a type of β-barrel outer membrane protein (OMP) called an autotransporter. The biogenesis of OMPs is typically mediated by the barrel assembly machinery (BAM) complex. In this study, we investigate the evolution, composition, and structure of the OMP biogenesis machinery in F. nucleatum. Our bioinformatics and proteomics analyses indicate that OMP biogenesis in F. nucleatum is mediated solely by the core component BamA. The structure of FnBamA highlights distinct features, including four POTRA domains and a C-terminal 16-stranded β-barrel domain observed as an inverted dimer. FnBamA represents the original composition of the assembly machinery, and a duplication event that resulted in BamA and TamA occurred after the split of other lineages, including the Proteobacteria, from the Fusobacteria. FnBamA, therefore, likely serves a singular role in the biogenesis of all OMPs.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
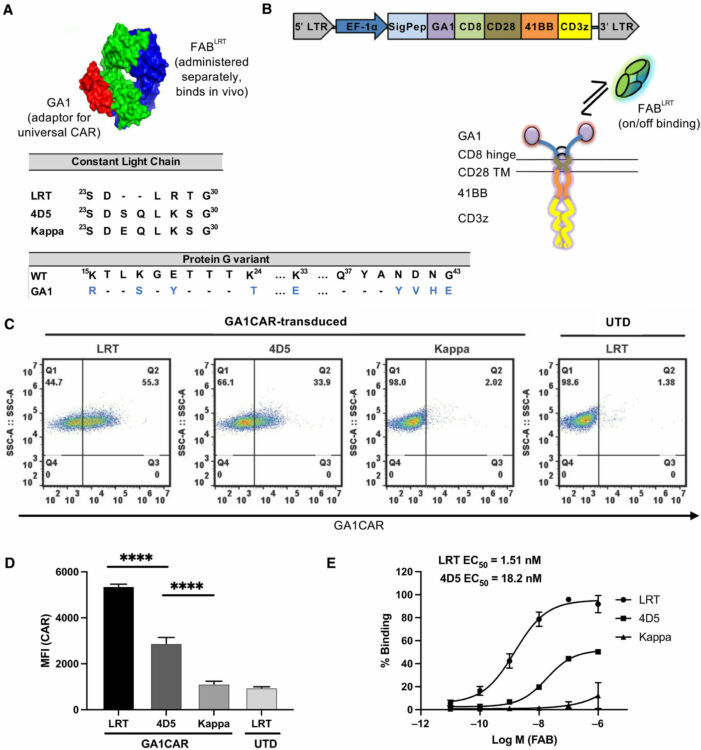
Arina, Ainhoa; Arauz, Edwin; Masoumi, Elham; Warzecha, Karolina W; Sääf, Annika; Widło, Łukasz; Slezak, Tomasz; Zieminska, Aleksandra; Dudek, Karolina; Schaefer, Zachary P; Lecka, Maria; Usatyuk, Svitlana; Weichselbaum, Ralph R; Kossiakoff, Anthony A
A universal chimeric antigen receptor (CAR)-fragment antibody binder (FAB) split system for cancer immunotherapy Journal Article
In: Sci Adv, vol. 11, no. 27, pp. eadv4937, 2025, ISSN: 2375-2548.
@article{pmid40614208,
title = {A universal chimeric antigen receptor (CAR)-fragment antibody binder (FAB) split system for cancer immunotherapy},
author = {Ainhoa Arina and Edwin Arauz and Elham Masoumi and Karolina W Warzecha and Annika Sääf and Łukasz Widło and Tomasz Slezak and Aleksandra Zieminska and Karolina Dudek and Zachary P Schaefer and Maria Lecka and Svitlana Usatyuk and Ralph R Weichselbaum and Anthony A Kossiakoff},
doi = {10.1126/sciadv.adv4937},
issn = {2375-2548},
year = {2025},
date = {2025-07-01},
urldate = {2025-07-01},
journal = {Sci Adv},
volume = {11},
number = {27},
pages = {eadv4937},
abstract = {Chimeric antigen receptor (CAR) T cell therapy has shown extraordinary results in treating hematological cancer but faces challenges like antigen loss, toxicity, and complex manufacturing. Universal and modular CAR constructs offer improved flexibility, safety, and cost-effectiveness over conventional CAR constructs. We present a CAR-fragment antibody binder (Fab) platform on the basis of an engineered protein G variant (GA1) and Fab scaffolds. Expression of GA1CAR on human CD8 T cells leads to antigen recognition and T cell effector function that can be modulated according to the affinity of the CAR for the Fab and of the Fab for the target. GA1CAR T cells can recognize multiple Fab-antigen pairs on breast and ovarian cancer cell lines. Adoptively transferred GA1CAR T cells control tumors in breast cancer xenograft models, and their targeting can be quickly redirected using different Fabs. This versatile "plug-and-play" CAR T platform has potential for application in personalized therapy, preventing antigen loss variant escape, decreasing toxicity, and increasing access.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
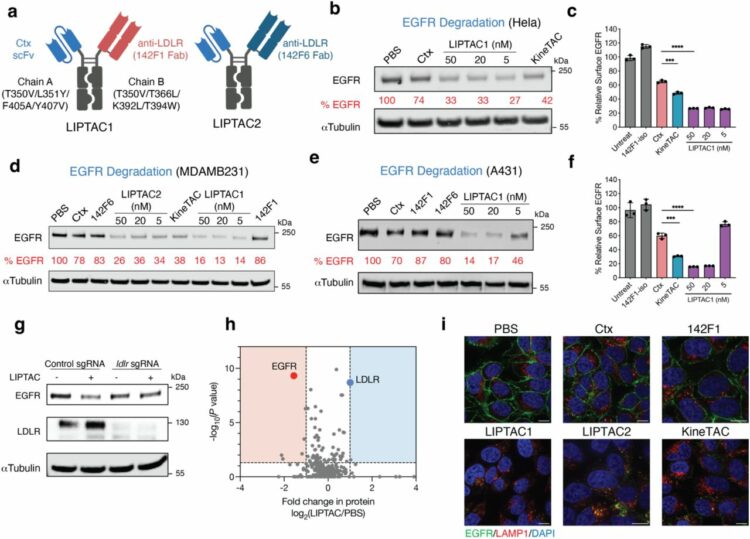
Zhao, Fangzhu; Wu, Yan; Schaefer, Kaitlin; Zhang, Yun; Miao, Kun; Yao, Zi; Ganjave, Snehal D; Kumru, Kaan; Peters-Clarke, Trenton M; Inague, Alex; Olzmann, James A; Leung, Kevin K; Wells, James A
Low-density lipoprotein receptor-targeting chimeras for membrane protein degradation and enhanced drug delivery Journal Article
In: bioRxiv, 2025, ISSN: 2692-8205.
@article{pmid40661577,
title = {Low-density lipoprotein receptor-targeting chimeras for membrane protein degradation and enhanced drug delivery},
author = {Fangzhu Zhao and Yan Wu and Kaitlin Schaefer and Yun Zhang and Kun Miao and Zi Yao and Snehal D Ganjave and Kaan Kumru and Trenton M Peters-Clarke and Alex Inague and James A Olzmann and Kevin K Leung and James A Wells},
doi = {10.1101/2025.06.06.658366},
issn = {2692-8205},
year = {2025},
date = {2025-06-01},
urldate = {2025-06-01},
journal = {bioRxiv},
abstract = {Antibody-based therapeutics encompass diverse modalities for targeting tumor cells. Among these, antibody-drug conjugates (ADCs) and extracellular targeted protein degradation (eTPD) specifically depend on efficient lysosomal trafficking for activity. However, many tumor antigens exhibit poor internalization, limiting ADC effectiveness. To address this, we developed low-density lipoprotein receptor-targeting chimeras (LIPTACs), leveraging the constitutive endocytic and recycling activity of the LDLR to enhance lysosomal delivery. LIPTACs enable efficient and selective degradation of diverse extracellular membrane proteins. Additionally, by coupling LIPTACs with cytotoxic payloads to generate degrader-drug conjugates, we can achieve superior intracellular delivery and enhanced cytotoxicity compared to conventional ADCs. The dual modality addresses key challenges of inadequate internalization in conventional ADCs and cytotoxic potency for current eTPD strategies. Our findings demonstrate that LDLR-mediated trafficking can enhance eTPD and ADCs, providing a hybrid blueprint for developing next-generation antibody therapeutics with broader utility and improved efficacy in cancer treatment.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
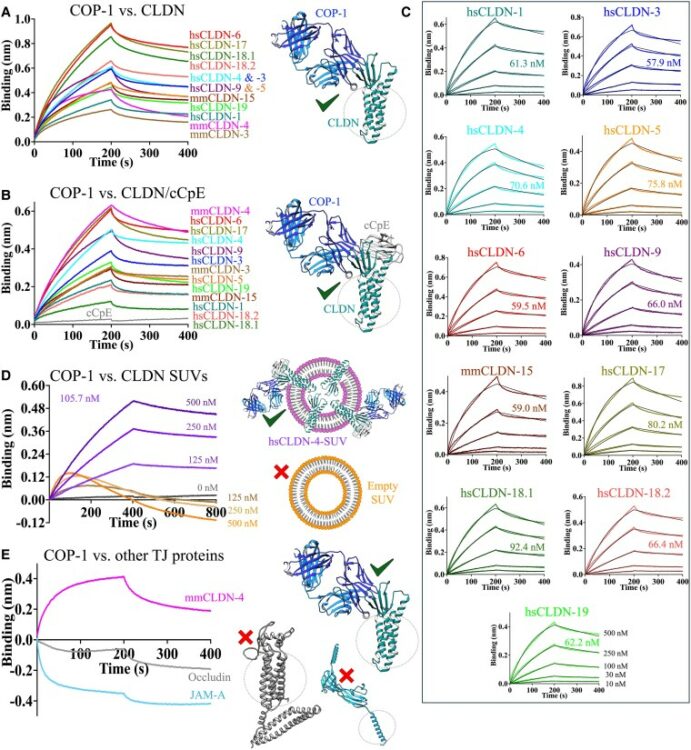
Ogbu, Chinemerem P; de Las Alas, Mason; Mandriota, Alexandria M; Liu, Xiangdong; Kapoor, Srajan; Choudhury, Jagrity; Ruma, Yasmeen N; Goodman, Michael C; Sanders, Charles R; Gonen, Tamir; Kossiakoff, Anthony A; Duffey, Michael E; Vecchio, Alex J
Biophysical basis of tight junction barrier modulation by a pan-claudin-binding molecule Journal Article
In: PNAS Nexus, vol. 4, no. 6, pp. pgaf189, 2025, ISSN: 2752-6542.
@article{pmid40575703,
title = {Biophysical basis of tight junction barrier modulation by a pan-claudin-binding molecule},
author = {Chinemerem P Ogbu and Mason de Las Alas and Alexandria M Mandriota and Xiangdong Liu and Srajan Kapoor and Jagrity Choudhury and Yasmeen N Ruma and Michael C Goodman and Charles R Sanders and Tamir Gonen and Anthony A Kossiakoff and Michael E Duffey and Alex J Vecchio},
doi = {10.1093/pnasnexus/pgaf189},
issn = {2752-6542},
year = {2025},
date = {2025-06-01},
urldate = {2025-06-01},
journal = {PNAS Nexus},
volume = {4},
number = {6},
pages = {pgaf189},
abstract = {Claudins are a 27-member family of membrane proteins that form and fortify specialized cell contacts in endothelium and epithelium called tight junctions. Tight junctions restrict paracellular transport through tissues by forming molecular barriers between cells. Claudin-binding molecules thus hold promise for modulating tight junction permeability to deliver drugs or as therapeutics to treat tight junction-linked disease. The development of claudin-binding molecules, however, is hindered by their physicochemical intractability and small targetable surfaces. Here, we determine that a synthetic antibody fragment (sFab) that we developed binds with nanomolar affinity directly to 10 claudin subtypes and other distantly related claudin family members but not to other tight junction-localized membrane proteins. It does so by targeting the extracellular surfaces of claudins, which we verify by applying this sFab to a model intestinal epithelium and observe that it opens paracellular barriers comparable to a known, but application limited, tight junction modulating protein. This pan-claudin-binding molecule holds potential for both basic and translational applications as it is a probe of claudin and tight junction structure in vitro and in vivo and a tool to modulate the permeability of tight junctions broadly across tissue barriers.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
O'Leary, Kelly M; Slezak, Tomasz; Kossiakoff, Anthony A
Conformation-specific synthetic intrabodies modulate mTOR signaling with subcellular spatial resolution Journal Article
In: Proc Natl Acad Sci U S A, vol. 122, no. 24, pp. e2424679122, 2025, ISSN: 1091-6490.
@article{pmid40489625,
title = {Conformation-specific synthetic intrabodies modulate mTOR signaling with subcellular spatial resolution},
author = {Kelly M O'Leary and Tomasz Slezak and Anthony A Kossiakoff},
doi = {10.1073/pnas.2424679122},
issn = {1091-6490},
year = {2025},
date = {2025-06-01},
urldate = {2025-06-01},
journal = {Proc Natl Acad Sci U S A},
volume = {122},
number = {24},
pages = {e2424679122},
abstract = {Subcellular compartmentalization is integral to the spatial regulation of mechanistic target of rapamycin (mTOR) signaling. However, the biological outputs associated with location-specific mTOR signaling events are poorly understood and challenging to decouple. Here, we engineered synthetic intracellular antibodies (intrabodies) that are capable of modulating mTOR signaling with genetically programmable spatial resolution. Epitope-directed phage display was exploited to generate high affinity synthetic antibody fragments (Fabs) against the FKBP12-Rapamycin binding site of mTOR (mTOR). We determined high-resolution crystal structures of two unique Fabs that discriminate distinct conformational states of mTOR through recognition of its substrate recruitment interface. By leveraging these conformation-specific binders as intracellular probes, we uncovered the structural basis for an allosteric mechanism governing mTOR complex 1 (mTORC1) stability mediated by subtle structural adjustments within mTOR. Furthermore, our results demonstrated that synthetic binders emulate natural substrates by employing divergent yet complementary hydrophobic residues at defined positions, underscoring the broad molecular recognition capability of mTOR. Intracellular signaling studies showed differential time-dependent inhibition of S6 kinase 1 and Akt phosphorylation by genetically encoded intrabodies, thus supporting a mechanism of inhibition analogous to the natural product rapamycin. Finally, we implemented a feasible approach to selectively modulate mTOR signaling in the nucleus through spatially programmed intrabody expression. These findings establish intrabodies as versatile tools for dissecting the conformational regulation of mTORC1 and should be useful to explore how location-specific mTOR signaling influences disease progression.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
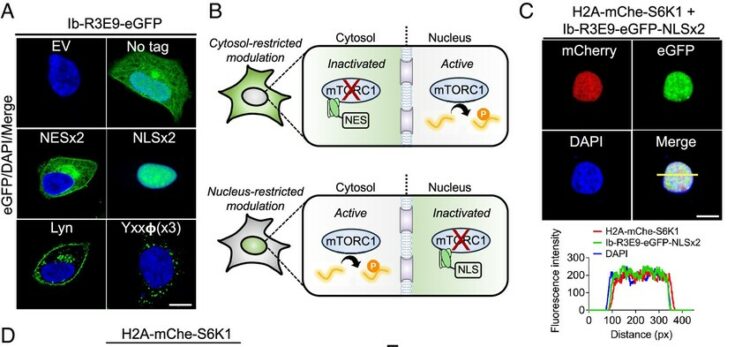
Adams, Jarrett J; Bruce, Heather A; Subramania, Suryasree; Ploder, Lynda; Garcia, Julia; Pot, Isabelle; Blazer, Levi L; Singer, Alexander U; Sidhu, Sachdev S
Synthetic antibodies targeting EphA2 induce diverse signaling-competent clusters with differential activation Journal Article
In: Protein Sci, vol. 34, no. 6, pp. e70145, 2025, ISSN: 1469-896X.
@article{pmid40411427,
title = {Synthetic antibodies targeting EphA2 induce diverse signaling-competent clusters with differential activation},
author = {Jarrett J Adams and Heather A Bruce and Suryasree Subramania and Lynda Ploder and Julia Garcia and Isabelle Pot and Levi L Blazer and Alexander U Singer and Sachdev S Sidhu},
doi = {10.1002/pro.70145},
issn = {1469-896X},
year = {2025},
date = {2025-06-01},
urldate = {2025-06-01},
journal = {Protein Sci},
volume = {34},
number = {6},
pages = {e70145},
abstract = {The receptor tyrosine kinase EphA2 interacts with ephrin (Efn) ligands to mediate bi-directional signals that drive cellular sorting processes during tissue development. In the context of various cancers, EphA2 can also drive invasive metastatic disease and represents an important target for cancer therapeutics. Natural Efn ligands sterically seed intertwined EphA2 clusters capable of recruiting intracellular kinases to mediate trans-phosphorylation. Synthetic proteins, such as antibodies (Abs), can mimic Efn ligands to trigger EphA2 signaling, leading to receptor internalization and degradation, and enabling intracellular delivery of conjugated drugs. Furthermore, Abs are capable of recruiting EphA2 into clusters distinct from those seeded by Efn. We developed three synthetic Abs targeting distinct EphA2 domains and determined the paratope valency necessary for agonist or antagonist properties of each of the three epitopes. Structural modeling of monovalent Fabs in complex with EphA2 elucidated competitive and non-competitive mechanisms of inhibition of EphA2 canonical signaling. Likewise, modeling of clusters induced by bivalent IgGs elucidated multiple signaling-competent EphA2 clusters capable of triggering a continuum of signaling strengths and provided insights into the requirement for multimerization of EphA2 to trigger phosphorylation. Our study shows how different agonist clusters lead to distinct kinase recruitment efficiencies to modify phosphotyrosine signal strength, and provides a panel of anti-EphA2 Abs as reagents for the development of therapeutics.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
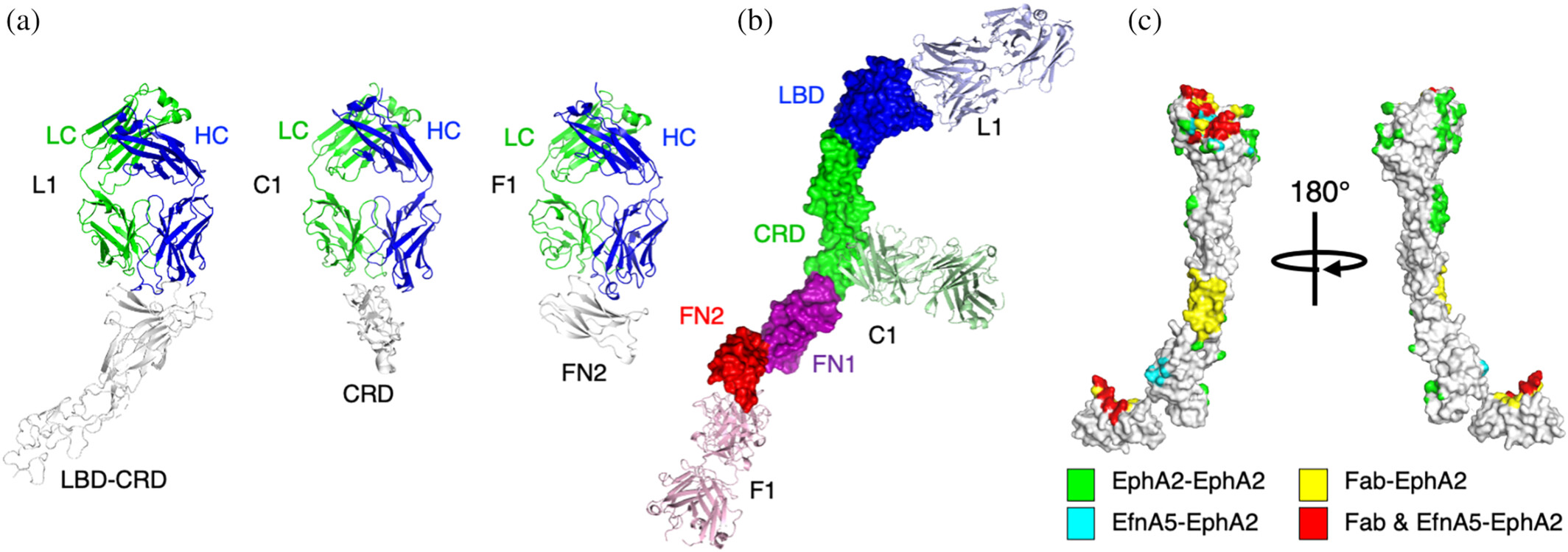
Miersch, Shane; Singer, Alex U; Chen, Chao; Fellouse, Fred; Gopalsamy, Anupriya; Costa, Luciano Silva E; Lacasta, Anna; Chege, Hannah; Chege, Naomi; Nene, Vishvanath; Sidhu, Sachdev S
Molecular characterization of a synthetic neutralizing antibody targeting p67 of Theileria parva Journal Article
In: Protein Sci, vol. 34, no. 6, pp. e70153, 2025, ISSN: 1469-896X.
@article{pmid40384600,
title = {Molecular characterization of a synthetic neutralizing antibody targeting p67 of Theileria parva},
author = {Shane Miersch and Alex U Singer and Chao Chen and Fred Fellouse and Anupriya Gopalsamy and Luciano Silva E Costa and Anna Lacasta and Hannah Chege and Naomi Chege and Vishvanath Nene and Sachdev S Sidhu},
doi = {10.1002/pro.70153},
issn = {1469-896X},
year = {2025},
date = {2025-06-01},
urldate = {2025-06-01},
journal = {Protein Sci},
volume = {34},
number = {6},
pages = {e70153},
abstract = {The Theileria parva sporozoite surface antigen p67 is a target of the bovine humoral immune response that generates antibodies capable of providing protection against subsequent infection. As a result, p67 has been the subject of efforts aimed at the development of an anti-sporozoite subunit vaccine. Previous studies have identified neutralizing epitopes in the N- and C-terminal regions of the full-length protein and shown that immunization with a C-terminal fragment of p67 (p67C) alone is capable of eliciting protection. To identify additional neutralizing epitopes in p67C, selections were conducted against it using a phage-displayed synthetic antibody library. An antibody that neutralized the sporozoite in vitro was identified, and the crystal structure of a Fab:peptide complex was elucidated. Mutagenesis studies aimed at validating and further characterizing the Fab:peptide interaction identified critical residues involved in binding and neutralization. This study also validates distinct epitopes for previously reported neutralizing antibodies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
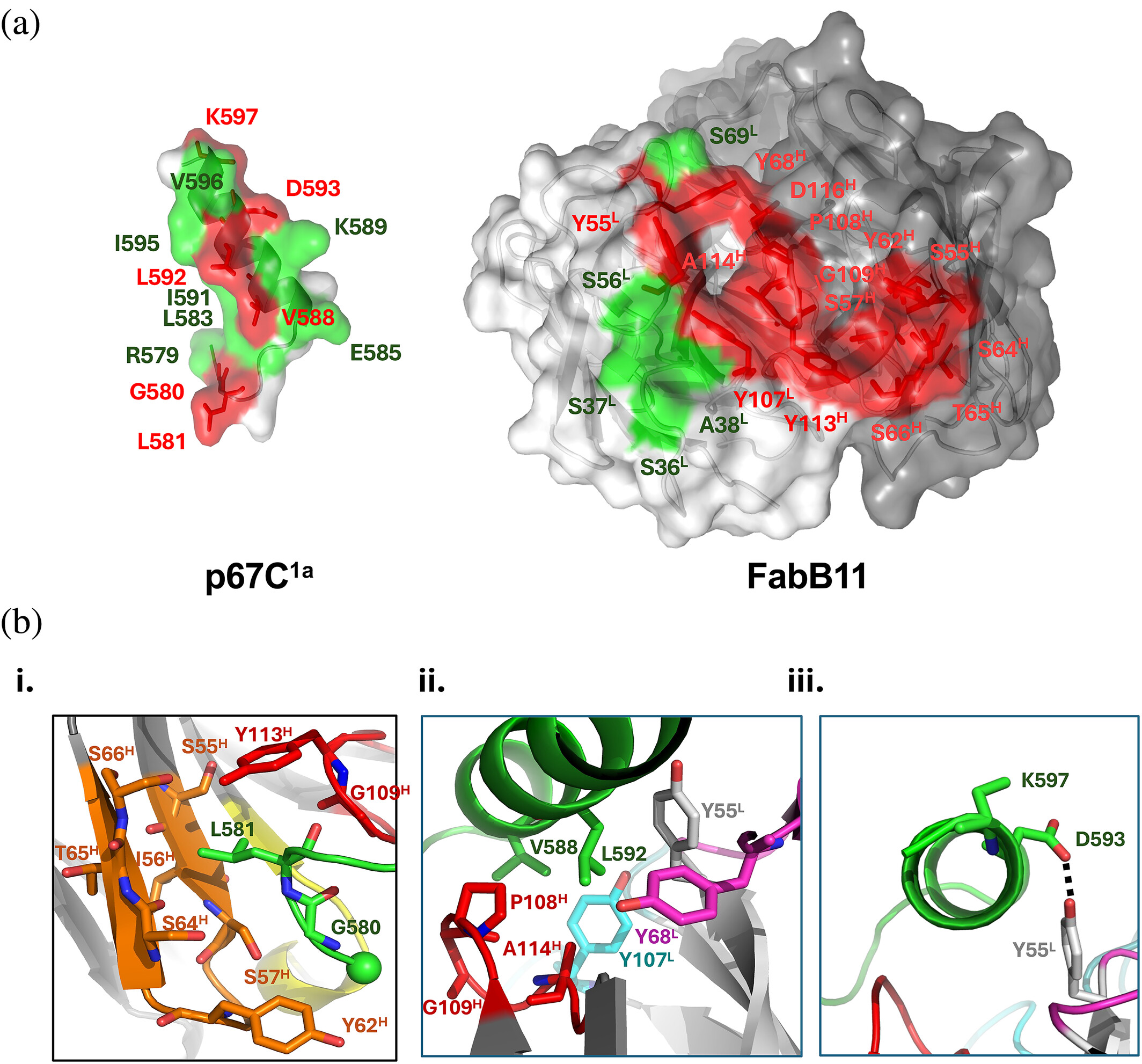
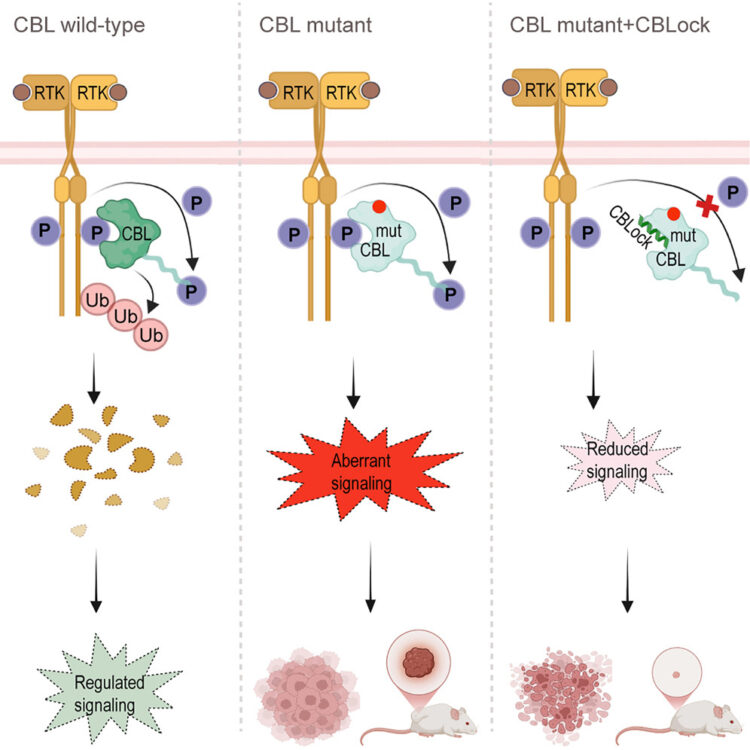
Ahmed, Syed Feroj; Anand, Jayanthi; Zhang, Wei; Buetow, Lori; Rishi, Loveena; Mitchell, Louise; Bohlen, Jonathan; Lilla, Sergio; Sibbet, Gary J; Nixon, Colin; Patel, Amrita; Majorek, Karolina A; Zanivan, Sara; Bustamante, Jacinta C; Sidhu, Sachdev S; Blyth, Karen; Huang, Danny T
Locking CBL TKBD in its native conformation presents a novel therapeutic opportunity in mutant CBL-dependent leukemia Journal Article
In: Mol Ther, 2025, ISSN: 1525-0024.
@article{pmid40329529,
title = {Locking CBL TKBD in its native conformation presents a novel therapeutic opportunity in mutant CBL-dependent leukemia},
author = {Syed Feroj Ahmed and Jayanthi Anand and Wei Zhang and Lori Buetow and Loveena Rishi and Louise Mitchell and Jonathan Bohlen and Sergio Lilla and Gary J Sibbet and Colin Nixon and Amrita Patel and Karolina A Majorek and Sara Zanivan and Jacinta C Bustamante and Sachdev S Sidhu and Karen Blyth and Danny T Huang},
doi = {10.1016/j.ymthe.2025.04.042},
issn = {1525-0024},
year = {2025},
date = {2025-05-01},
urldate = {2025-05-01},
journal = {Mol Ther},
abstract = {Casitas B-lineage lymphoma (CBL) is an E3 ubiquitin ligase critical for negatively regulating receptor protein tyrosine kinases (RTKs). Deleterious CBL mutants lose E3 activity, but act as adaptors that gain function to cause myeloproliferative neoplasms. Currently, there is no targeted treatment available for patients with CBL mutant-dependent disorders. By combining phage-display technology and structure-based optimization, we discovered CBLock, a nanomolar affinity peptide inhibitor, that binds the substrate-binding site of CBL's tyrosine kinase binding domain (TKBD). CBLock disrupts the interaction between CBL mutants and RTKs, thereby impairing RTK-mediated priming of adaptor function of CBL mutants and downstream signaling. Notably, CBLock binds TKBD without inducing conformational changes, thereby preserving its ligand-free native conformation. In contrast, when CBL binds RTK substrates, TKBD undergoes a conformational change. Maintaining the native CBL TKBD conformation was crucial for CBLock to inhibit proliferation, induce cell-cycle arrest, and promote apoptosis in leukemia cells harboring CBL mutations. In a mouse xenograft model of acute myeloid leukemia (AML), CBLock reduced tumor burden and improved survival rate. Moreover, CBLock inhibited the proliferation of cells derived from patients with CBL mutations. Therefore, inhibiting CBL TKBD in its native state presents a promising therapeutic opportunity in targeting mutant CBL-dependent leukemia.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Qualls, Anita E; Tsao, Tasha; Lui, Irene; Lim, Shion A; Su, Yapeng; Chen, Ernie; Duchen, Dylan; Maecker, Holden T; Kim-Schulze, Seunghee; Montgomery, Ruth R; Krammer, Florian; Langelier, Charles R; Levy, Ofer; Baden, Lindsey R; Melamed, Esther; Ehrlich, Lauren Ir; McComsey, Grace A; Sekaly, Rafick P; Cairns, Charles B; Haddad, Elias K; Shaw, Albert C; Hafler, David A; Corry, David B; Kheradmand, Farrah; Atkinson, Mark A; Brakenridge, Scott C; Higuita, Nelson I Agudelo; Metcalf, Jordan P; Hough, Catherine L; Messer, William B; Pulendran, Bali; Nadeau, Kari C; Davis, Mark M; Fernandez-Sesma, Ana; Simon, Viviana; Kraft, Monica; Bime, Christian; Calfee, Carolyn S; Erle, David J; Schaenmann, Joanna; Ozonoff, Al; Peters, Bjoern; Kleinstein, Steven H; Augustine, Alison D; Diray-Arce, Joann; Becker, Patrice M; Rouphael, Nadine; Network, Impacc; Goldman, Jason D; Calabrese, Daniel R; Heath, James R; Wells, James A; Reed, Elaine F; Lanier, Lewis L; Pickering, Harry; Aguilar, Oscar A
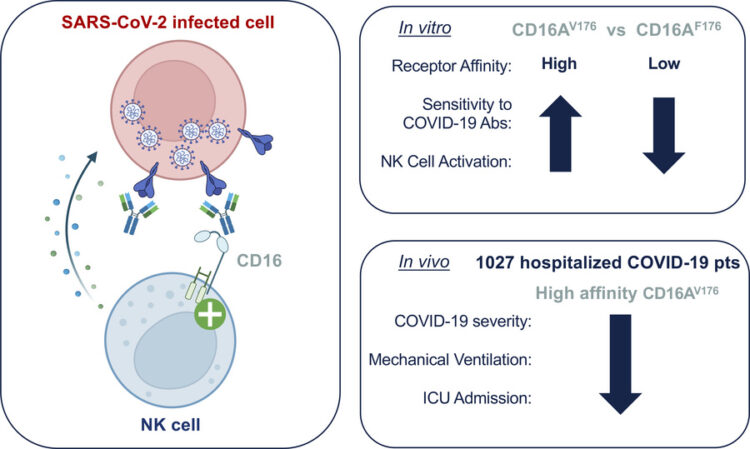
High affinity CD16 polymorphism associated with reduced risk of severe COVID-19 Journal Article
In: JCI Insight, 2025, ISSN: 2379-3708.
@article{pmid40402577,
title = {High affinity CD16 polymorphism associated with reduced risk of severe COVID-19},
author = {Anita E Qualls and Tasha Tsao and Irene Lui and Shion A Lim and Yapeng Su and Ernie Chen and Dylan Duchen and Holden T Maecker and Seunghee Kim-Schulze and Ruth R Montgomery and Florian Krammer and Charles R Langelier and Ofer Levy and Lindsey R Baden and Esther Melamed and Lauren Ir Ehrlich and Grace A McComsey and Rafick P Sekaly and Charles B Cairns and Elias K Haddad and Albert C Shaw and David A Hafler and David B Corry and Farrah Kheradmand and Mark A Atkinson and Scott C Brakenridge and Nelson I Agudelo Higuita and Jordan P Metcalf and Catherine L Hough and William B Messer and Bali Pulendran and Kari C Nadeau and Mark M Davis and Ana Fernandez-Sesma and Viviana Simon and Monica Kraft and Christian Bime and Carolyn S Calfee and David J Erle and Joanna Schaenmann and Al Ozonoff and Bjoern Peters and Steven H Kleinstein and Alison D Augustine and Joann Diray-Arce and Patrice M Becker and Nadine Rouphael and Impacc Network and Jason D Goldman and Daniel R Calabrese and James R Heath and James A Wells and Elaine F Reed and Lewis L Lanier and Harry Pickering and Oscar A Aguilar},
doi = {10.1172/jci.insight.191314},
issn = {2379-3708},
year = {2025},
date = {2025-05-01},
urldate = {2025-05-01},
journal = {JCI Insight},
abstract = {CD16 is an activating Fc receptor on natural killer cells that mediates antibody-dependent cellular cytotoxicity (ADCC), a key mechanism in antiviral immunity. However, the role of NK cell-mediated ADCC in SARS-CoV-2 infection remains unclear, particularly whether it limits viral spread and disease severity or contributes to the immunopathogenesis of COVID-19. We hypothesized that the high-affinity CD16AV176 polymorphism influences these outcomes. Using a novel in vitro reporter system, we demonstrated that CD16AV176 is a more potent and sensitive activator than the common CD16AF176 allele. To assess its clinical relevance, we analyzed 1,027 hospitalized COVID-19 patients from the Immunophenotyping Assessment in a COVID-19 Cohort (IMPACC), a comprehensive longitudinal dataset with extensive transcriptomic, proteomic, and clinical data. The high-affinity CD16AV176 allele was associated with a significantly reduced risk of ICU admission, mechanical ventilation, and severe disease trajectories. Lower anti-SARS-CoV-2 IgG titers were correlated to CD16AV176; however, there was no difference in viral load across CD16 genotypes. Proteomic analysis revealed that participants homozygous for CD16AV176 had lower levels of inflammatory mediators. These findings suggest that CD16AV176 enhances early NK cell-mediated immune responses, limiting severe respiratory complications in COVID-19. This study identifies a protective genetic factor against severe COVID-19, informing future host-directed therapeutic strategies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
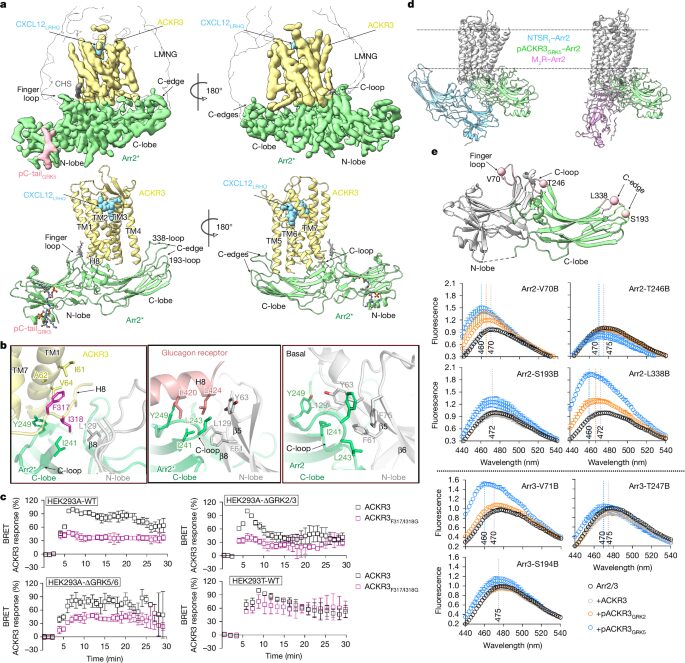
Chen, Qiuyan; Schafer, Christopher T; Mukherjee, Somnath; Wang, Kai; Gustavsson, Martin; Fuller, James R; Tepper, Katelyn; Lamme, Thomas D; Aydin, Yasmin; Agrawal, Parth; Terashi, Genki; Yao, Xin-Qiu; Kihara, Daisuke; Kossiakoff, Anthony A; Handel, Tracy M; Tesmer, John J G
Effect of phosphorylation barcodes on arrestin binding to a chemokine receptor Journal Article
In: Nature, 2025, ISSN: 1476-4687.
@article{pmid40399676,
title = {Effect of phosphorylation barcodes on arrestin binding to a chemokine receptor},
author = {Qiuyan Chen and Christopher T Schafer and Somnath Mukherjee and Kai Wang and Martin Gustavsson and James R Fuller and Katelyn Tepper and Thomas D Lamme and Yasmin Aydin and Parth Agrawal and Genki Terashi and Xin-Qiu Yao and Daisuke Kihara and Anthony A Kossiakoff and Tracy M Handel and John J G Tesmer},
doi = {10.1038/s41586-025-09024-9},
issn = {1476-4687},
year = {2025},
date = {2025-05-01},
urldate = {2025-05-01},
journal = {Nature},
abstract = {Unique phosphorylation 'barcodes' installed in different regions of an active seven-transmembrane receptor by different G-protein-coupled receptor (GPCR) kinases (GRKs) have been proposed to promote distinct cellular outcomes, but it is unclear whether or how arrestins differentially engage these barcodes. Here, to address this, we developed an antigen-binding fragment (Fab7) that recognizes both active arrestin2 (β-arrestin1) and arrestin3 (β-arrestin2) without interacting with bound receptor polypeptides. We used Fab7 to determine the structures of both arrestins in complex with atypical chemokine receptor 3 (ACKR3) phosphorylated in different regions of its C-terminal tail by either GRK2 or GRK5 (ref. ). The GRK2-phosphorylated ACKR3 resulted in more heterogeneous 'tail-mode' assemblies, whereas phosphorylation by GRK5 resulted in more rigid 'ACKR3-adjacent' assemblies. Unexpectedly, the finger loops of both arrestins engaged the micelle surface rather than the receptor intracellular pocket, with arrestin3 being more dynamic, partly because of its lack of a membrane-anchoring motif. Thus, both the region of the barcode and the arrestin isoform involved can alter the structure and dynamics of GPCR-arrestin complexes, providing a possible mechanistic basis for unique downstream cellular effects, such as the efficiency of chemokine scavenging and the robustness of arrestin binding in ACKR3.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Lui, Irene; Schaefer, Kaitlin; Kirkemo, Lisa L; Zhou, Jie; Perera, Rushika M; Leung, Kevin K; Wells, James A
Hypoxia Induces Extensive Protein and Proteolytic Remodeling of the Cell Surface in Pancreatic Adenocarcinoma (PDAC) Cell Lines Journal Article
In: J Proteome Res, 2025, ISSN: 1535-3907.
@article{pmid40312771,
title = {Hypoxia Induces Extensive Protein and Proteolytic Remodeling of the Cell Surface in Pancreatic Adenocarcinoma (PDAC) Cell Lines},
author = {Irene Lui and Kaitlin Schaefer and Lisa L Kirkemo and Jie Zhou and Rushika M Perera and Kevin K Leung and James A Wells},
doi = {10.1021/acs.jproteome.4c01037},
issn = {1535-3907},
year = {2025},
date = {2025-05-01},
urldate = {2025-05-01},
journal = {J Proteome Res},
abstract = {The tumor microenvironment (TME) plays a crucial role in cancer progression. Hypoxia is a hallmark of the TME and induces a cascade of molecular events that affect cellular processes involved in metabolism, metastasis, and proteolysis. In pancreatic ductal adenocarcinoma (PDAC), tumor tissues are extremely hypoxic. Here, we leveraged mass spectrometry technologies to examine hypoxia-induced alterations in the abundance and proteolytic modifications to cell surface and secreted proteins. Across four PDAC cell lines, we discovered extensive proteolytic remodeling of cell surface proteins involved in cellular adhesion and motility. Looking outward at the surrounding secreted space, we identified hypoxia-regulated secreted and proteolytically shed proteins involved in regulating the humoral immune and inflammatory response, and an upregulation of proteins involved in metabolic processing and tissue development. Combining cell surface N-terminomics and secretomics to evaluate the cellular response to hypoxia enabled us to identify significantly altered candidate proteins which may serve as potential biomarkers and therapeutic targets in PDAC. Furthermore, this approach provides a blueprint for studying dysregulated extracellular proteolysis in other cancers and inflammatory diseases.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
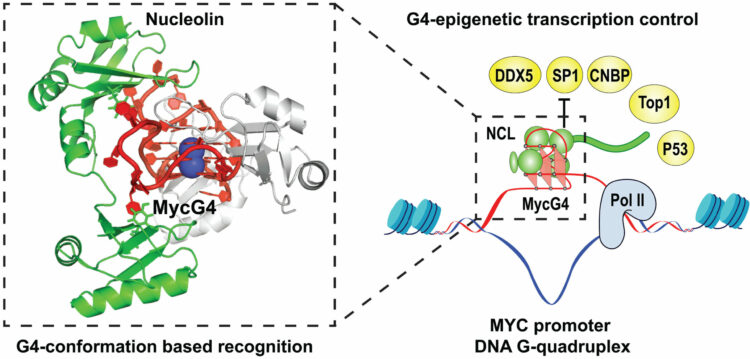
Chen, Luying; Dickerhoff, Jonathan; Zheng, Ke-wei; Erramilli, Satchal; Feng, Hanqiao; Wu, Guanhui; Onel, Buket; Chen, Yuwei; Wang, Kai-Bo; Carver, Megan; Lin, Clement; Sakai, Saburo; Wan, Jun; Vinson, Charles; Hurley, Laurence; Kossiakoff, Anthony A.; Deng, Nanjie; Bai, Yawen; Noinaj, Nicholas; Yang, Danzhou
Structural basis for nucleolin recognition of MYC promoter G-quadruplex Journal Article
In: Science, vol. 388, no. 6744, 2025, ISSN: 1095-9203.
@article{Chen2025,
title = {Structural basis for nucleolin recognition of MYC promoter G-quadruplex},
author = {Luying Chen and Jonathan Dickerhoff and Ke-wei Zheng and Satchal Erramilli and Hanqiao Feng and Guanhui Wu and Buket Onel and Yuwei Chen and Kai-Bo Wang and Megan Carver and Clement Lin and Saburo Sakai and Jun Wan and Charles Vinson and Laurence Hurley and Anthony A. Kossiakoff and Nanjie Deng and Yawen Bai and Nicholas Noinaj and Danzhou Yang},
doi = {10.1126/science.adr1752},
issn = {1095-9203},
year = {2025},
date = {2025-04-18},
urldate = {2025-04-18},
journal = {Science},
volume = {388},
number = {6744},
publisher = {American Association for the Advancement of Science (AAAS)},
abstract = {The MYC oncogene promoter G-quadruplex (MycG4) regulates transcription and is a prevalent G4 locus in immortal cells. Nucleolin, a major MycG4-binding protein, exhibits greater affinity for MycG4 than for nucleolin recognition element (NRE) RNA. Nucleolin’s four RNA binding domains (RBDs) are essential for high-affinity MycG4 binding. We present the 2.6-angstrom crystal structure of the nucleolin-MycG4 complex, revealing a folded parallel three-tetrad G-quadruplex with two coordinating potassium ions (K+), interacting with RBD1, RBD2, and Linker12 through its 6–nucleotide (nt) central loop and 5′ flanking region. RBD3 and RBD4 bind MycG4’s 1-nt loops as demonstrated by nuclear magnetic resonance (NMR). Cleavage under targets and tagmentation sequencing confirmed nucleolin’s binding to MycG4 in cells. Our results revealed a G4 conformation-based recognition by a regulating protein through multivalent interactions, suggesting that G4s are nucleolin’s primary cellular substrates, indicating G4 epigenetic transcriptional regulation and helping G4-targeted drug discovery.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
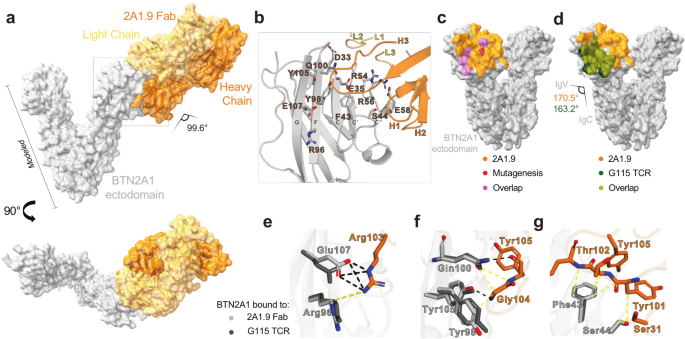
Ramesh, Amrita; Roy, Sobhan; Slezak, Tomasz; Fuller, James; Graves, Hortencia; Mamedov, Murad R.; Marson, Alexander; Kossiakoff, Anthony A.; Adams, Erin J.
Mapping the extracellular molecular architecture of the pAg-signaling complex with α-Butyrophilin antibodies Journal Article
In: Sci Rep, vol. 15, no. 1, 2025, ISSN: 2045-2322.
@article{Ramesh2025,
title = {Mapping the extracellular molecular architecture of the pAg-signaling complex with α-Butyrophilin antibodies},
author = {Amrita Ramesh and Sobhan Roy and Tomasz Slezak and James Fuller and Hortencia Graves and Murad R. Mamedov and Alexander Marson and Anthony A. Kossiakoff and Erin J. Adams},
doi = {10.1038/s41598-025-94347-w},
issn = {2045-2322},
year = {2025},
date = {2025-04-09},
urldate = {2025-12-00},
journal = {Sci Rep},
volume = {15},
number = {1},
publisher = {Springer Science and Business Media LLC},
abstract = {Target cells trigger Vγ9Vδ2 T cell activation by signaling the intracellular accumulation of phospho-antigen metabolites (pAgs) through Butyrophilin (BTN)-3A1 and BTN2A1 to the Vγ9Vδ2 T cell receptor (TCR). An incomplete understanding of the molecular dynamics in this signaling complex hampers Vγ9Vδ2 T cell immunotherapeutic efficacy. A panel of engineered α-BTN3A1 and α-BTN2A1 antibody (mAb) reagents was used to probe the roles of BTN3A1 and BTN2A1 in pAg signaling. Modified α-BTN3A1 mAbs with increased inter-Fab distances establish that tight clustering of BTN3A1 is not necessary to stimulate Vγ9Vδ2 T cell activation, and that antagonism may occur through occlusion of a critical binding interaction between BTN3A1 and a yet unknown co-receptor. Finally, a panel of additional α-BTN2A1 antagonists utilize different biophysical mechanisms to compete with Vγ9Vδ2 TCRs for BTN2A1 binding. The complex structures of BTN2A1 ectodomain and Fabs from three antagonist mAbs provide molecular insights into BTN2A1 epitopes critical for pAg-signaling.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
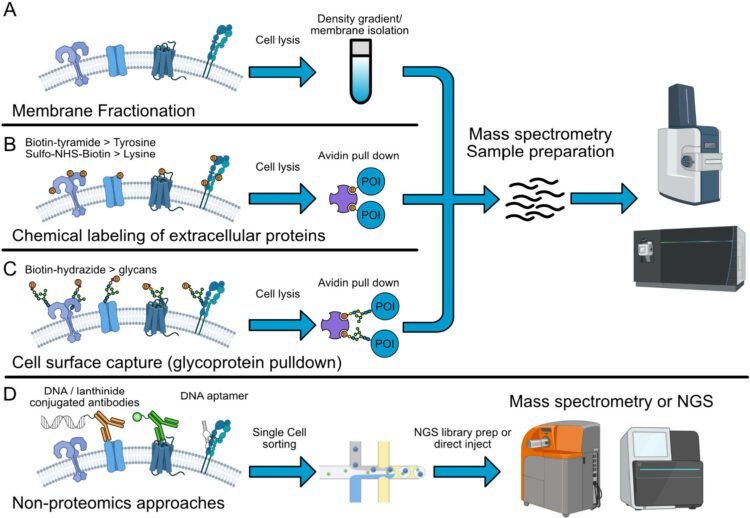
Leung, Kevin K; Schaefer, Kaitlin; Lin, Zhi; Yao, Zi; Wells, James A
Engineered Proteins and Chemical Tools to Probe the Cell Surface Proteome Journal Article
In: Chem Rev, 2025, ISSN: 1520-6890.
@article{pmid40178992,
title = {Engineered Proteins and Chemical Tools to Probe the Cell Surface Proteome},
author = {Kevin K Leung and Kaitlin Schaefer and Zhi Lin and Zi Yao and James A Wells},
doi = {10.1021/acs.chemrev.4c00554},
issn = {1520-6890},
year = {2025},
date = {2025-04-01},
urldate = {2025-04-01},
journal = {Chem Rev},
abstract = {The cell surface proteome, or surfaceome, is the hub for cells to interact and communicate with the outside world. Many disease-associated changes are hard-wired within the surfaceome, yet approved drugs target less than 50 cell surface proteins. In the past decade, the proteomics community has made significant strides in developing new technologies tailored for studying the surfaceome in all its complexity. In this review, we first dive into the unique characteristics and functions of the surfaceome, emphasizing the necessity for specialized labeling, enrichment, and proteomic approaches. An overview of surfaceomics methods is provided, detailing techniques to measure changes in protein expression and how this leads to novel target discovery. Next, we highlight advances in proximity labeling proteomics (PLP), showcasing how various enzymatic and photoaffinity proximity labeling techniques can map protein-protein interactions and membrane protein complexes on the cell surface. We then review the role of extracellular post-translational modifications, focusing on cell surface glycosylation, proteolytic remodeling, and the secretome. Finally, we discuss methods for identifying tumor-specific peptide MHC complexes and how they have shaped therapeutic development. This emerging field of neo-protein epitopes is constantly evolving, where targets are identified at the proteome level and encompass defined disease-associated PTMs, complexes, and dysregulated cellular and tissue locations. Given the functional importance of the surfaceome for biology and therapy, we view surfaceomics as a critical piece of this quest for neo-epitope target discovery.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
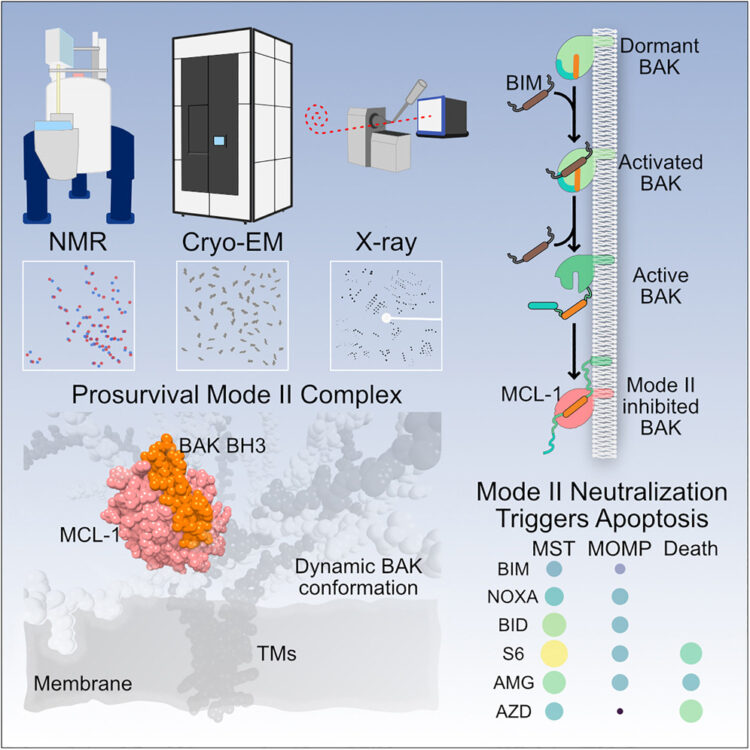
Srivastava, Shagun; Sekar, Giridhar; Ojoawo, Adedolapo; Aggarwal, Anup; Ferreira, Elisabeth; Uchikawa, Emiko; Yang, Meek; Grace, Christy R; Dey, Raja; Lin, Yi-Lun; Guibao, Cristina D; Jayaraman, Seetharaman; Mukherjee, Somnath; Kossiakoff, Anthony A; Dong, Bin; Myasnikov, Alexander; Moldoveanu, Tudor
Structural basis of BAK sequestration by MCL-1 in apoptosis Journal Article
In: Mol Cell, 2025, ISSN: 1097-4164.
@article{pmid40187349,
title = {Structural basis of BAK sequestration by MCL-1 in apoptosis},
author = {Shagun Srivastava and Giridhar Sekar and Adedolapo Ojoawo and Anup Aggarwal and Elisabeth Ferreira and Emiko Uchikawa and Meek Yang and Christy R Grace and Raja Dey and Yi-Lun Lin and Cristina D Guibao and Seetharaman Jayaraman and Somnath Mukherjee and Anthony A Kossiakoff and Bin Dong and Alexander Myasnikov and Tudor Moldoveanu},
doi = {10.1016/j.molcel.2025.03.013},
issn = {1097-4164},
year = {2025},
date = {2025-03-01},
urldate = {2025-03-01},
journal = {Mol Cell},
abstract = {Apoptosis controls cell fate, ensuring tissue homeostasis and promoting disease when dysregulated. The rate-limiting step in apoptosis is mitochondrial poration by the effector B cell lymphoma 2 (BCL-2) family proteins BAK and BAX, which are activated by initiator BCL-2 homology 3 (BH3)-only proteins (e.g., BIM) and inhibited by guardian BCL-2 family proteins (e.g., MCL-1). We integrated structural, biochemical, and pharmacological approaches to characterize the human prosurvival MCL-1:BAK complex assembled from their BCL-2 globular core domains. We reveal a canonical interaction with BAK BH3 bound to the hydrophobic groove of MCL-1 and disordered and highly dynamic BAK regions outside the complex interface. We predict similar conformations of activated effectors in complex with other guardians or effectors. The MCL-1:BAK complex is a major cancer drug target. We show that MCL-1 inhibitors are inefficient in neutralizing the MCL-1:BAK complex, requiring high doses to initiate apoptosis. Our study underscores the need to design superior clinical candidate MCL-1 inhibitors.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
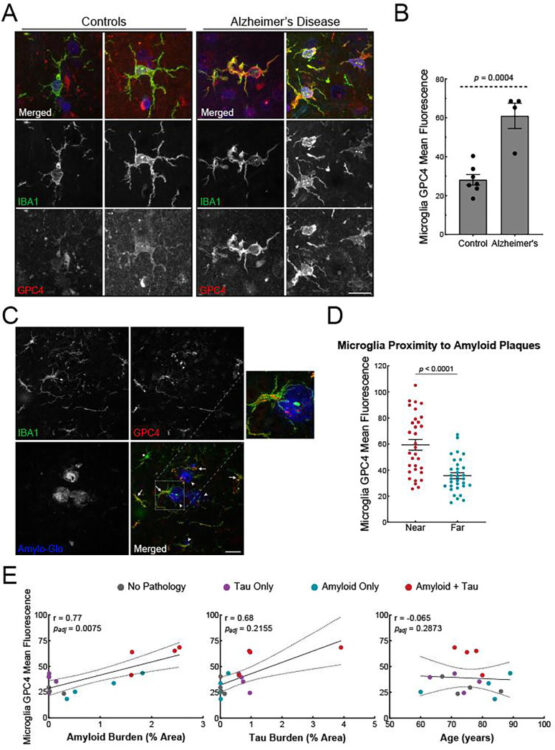
Holmes, Brandon B; Weigel, Thaddeus K; Chung, Jesseca M; Kaufman, Sarah K; Apresa, Brandon I; Byrnes, James R; Kumru, Kaan S; Vaquer-Alicea, Jaime; Gupta, Ankit; Rose, Indigo V L; Zhang, Yun; Nana, Alissa L; Alter, Dina; Grinberg, Lea T; Spina, Salvatore; Leung, Kevin K; Condello, Carlo; Kampmann, Martin; Seeley, William W; Coutinho-Budd, Jaeda C; Wells, James A
β-Amyloid Induces Microglial Expression of GPC4 and APOE Leading to Increased Neuronal Tau Pathology and Toxicity Journal Article
In: bioRxiv, 2025, ISSN: 2692-8205.
@article{pmid40060520,
title = {β-Amyloid Induces Microglial Expression of GPC4 and APOE Leading to Increased Neuronal Tau Pathology and Toxicity},
author = {Brandon B Holmes and Thaddeus K Weigel and Jesseca M Chung and Sarah K Kaufman and Brandon I Apresa and James R Byrnes and Kaan S Kumru and Jaime Vaquer-Alicea and Ankit Gupta and Indigo V L Rose and Yun Zhang and Alissa L Nana and Dina Alter and Lea T Grinberg and Salvatore Spina and Kevin K Leung and Carlo Condello and Martin Kampmann and William W Seeley and Jaeda C Coutinho-Budd and James A Wells},
doi = {10.1101/2025.02.20.637701},
issn = {2692-8205},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {bioRxiv},
abstract = {To elucidate the impact of Aβ pathology on microglia in Alzheimer's disease pathogenesis, we profiled the microglia surfaceome following treatment with Aβ fibrils. Our findings reveal that Aβ-associated human microglia upregulate Glypican 4 (GPC4), a GPI-anchored heparan sulfate proteoglycan (HSPG). In a amyloidosis model, glial GPC4 expression exacerbates motor deficits and reduces lifespan, indicating that glial GPC4 contributes to a toxic cellular program during neurodegeneration. In cell culture, GPC4 enhances microglia phagocytosis of tau aggregates, and shed GPC4 can act to facilitate tau aggregate uptake and seeding in neurons. Additionally, our data demonstrate that GPC4-mediated effects are amplified in the presence of APOE. These studies offer a mechanistic framework linking Aβ and tau pathology through microglial HSPGs and APOE.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Mann, Samuel I; Lin, Zhi; Tan, Sophia K; Zhu, Jiaqi; Widel, Zachary X W; Bakanas, Ian; Mansergh, Jarrett P; Liu, Rui; Kelly, Mark J S; Wu, Yibing; Wells, James A; Therien, Michael J; DeGrado, William F
De Novo Design of Proteins That Bind Naphthalenediimides, Powerful Photooxidants with Tunable Photophysical Properties Journal Article
In: J Am Chem Soc, 2025, ISSN: 1520-5126.
@article{pmid39982408,
title = {De Novo Design of Proteins That Bind Naphthalenediimides, Powerful Photooxidants with Tunable Photophysical Properties},
author = {Samuel I Mann and Zhi Lin and Sophia K Tan and Jiaqi Zhu and Zachary X W Widel and Ian Bakanas and Jarrett P Mansergh and Rui Liu and Mark J S Kelly and Yibing Wu and James A Wells and Michael J Therien and William F DeGrado},
doi = {10.1021/jacs.4c18151},
issn = {1520-5126},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {J Am Chem Soc},
abstract = { protein design provides a framework to test our understanding of protein function and build proteins with cofactors and functions not found in nature. Here, we report the design of proteins designed to bind powerful photooxidants and the evaluation of the use of these proteins to generate diffusible small-molecule reactive species. Because excited-state dynamics are influenced by the dynamics and hydration of a photooxidant's environment, it was important to not only design a binding site but also to evaluate its dynamic properties. Thus, we used computational design in conjunction with molecular dynamics (MD) simulations to design a protein, designated NBP (DI inding rotein), that held a naphthalenediimide (NDI), a powerful photooxidant, in a programmable molecular environment. Solution NMR confirmed the structure of the complex. We evaluated two NDI cofactors in this protein using ultrafast pump-probe spectroscopy to evaluate light-triggered intra- and intermolecular electron transfer function. Moreover, we demonstrated the utility of this platform to activate multiple molecular probes for protein labeling.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Gorelik, Maryna; Miersch, Shane; Sidhu, Sachdev S
Structural Survey of Antigen Recognition by Synthetic Human Antibodies Journal Article
In: Cold Spring Harb Protoc, vol. 2025, no. 2, pp. pdb.over107759, 2025, ISSN: 1559-6095.
@article{pmid38594044,
title = {Structural Survey of Antigen Recognition by Synthetic Human Antibodies},
author = {Maryna Gorelik and Shane Miersch and Sachdev S Sidhu},
doi = {10.1101/pdb.over107759},
issn = {1559-6095},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {Cold Spring Harb Protoc},
volume = {2025},
number = {2},
pages = {pdb.over107759},
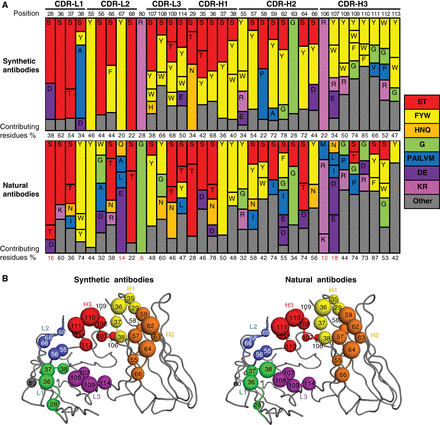
abstract = {Synthetic antibody libraries have been used extensively to isolate and optimize antibodies. To generate these libraries, the immunological diversity and the antibody framework(s) that supports it outside of the binding regions are carefully designed/chosen to ensure favorable functional and biophysical properties. In particular, minimalist, single-framework synthetic libraries pioneered by our group have yielded a vast trove of antibodies to a broad array of antigens. Here, we review their systematic and iterative development to provide insights into the design principles that make them a powerful tool for drug discovery. In addition, the ongoing accumulation of crystal structures of antigen-binding fragment (Fab)-antigen complexes generated with synthetic antibodies enables a deepening understanding of the structural determinants of antigen recognition and usage of immunoglobulin sequence diversity, which can assist in developing new strategies for antibody and library optimization. Toward this, we also survey here the structural landscape of a comprehensive and unbiased set of 50 distinct complexes derived from these libraries and compare it to a similar set of natural antibodies with the goal of better understanding how each achieves molecular recognition and whether opportunities exist for iterative improvement of synthetic libraries. From this survey, we conclude that despite the minimalist strategies used for design of these synthetic antibody libraries, the overall structural interaction landscapes are highly similar to natural repertoires. We also found, however, some key differences that can help guide the iterative design of new synthetic libraries via the introduction of positionally tailored diversity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Slezak, Tomasz; O'Leary, Kelly M; Li, Jinyang; Rohaim, Ahmed; Davydova, Elena K; Kossiakoff, Anthony A
Engineered protein G variants for multifunctional antibody-based assemblies Journal Article
In: Protein Sci, vol. 34, no. 2, pp. e70019, 2025, ISSN: 1469-896X.
@article{pmid39865354,
title = {Engineered protein G variants for multifunctional antibody-based assemblies},
author = {Tomasz Slezak and Kelly M O'Leary and Jinyang Li and Ahmed Rohaim and Elena K Davydova and Anthony A Kossiakoff},
doi = {10.1002/pro.70019},
issn = {1469-896X},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {Protein Sci},
volume = {34},
number = {2},
pages = {e70019},
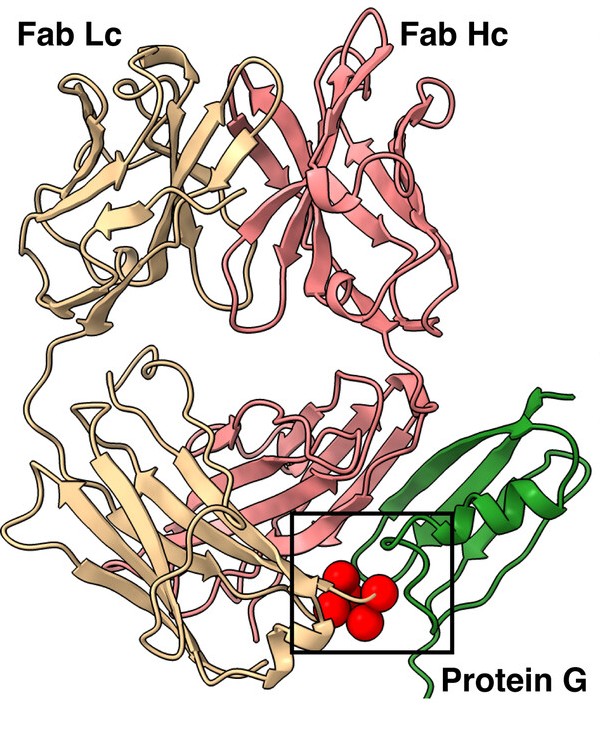
abstract = {We have developed a portfolio of antibody-based modules that can be prefabricated as standalone units and snapped together in plug-and-play fashion to create uniquely powerful multifunctional assemblies. The basic building blocks are derived from multiple pairs of native and modified Fab scaffolds and protein G (PG) variants engineered by phage display to introduce high pair-wise specificity. The variety of possible Fab-PG pairings provides a highly orthogonal system that can be exploited to perform challenging cell biology operations in a straightforward manner. The simplest manifestation allows multiplexed antigen detection using PG variants fused to fluorescently labeled SNAP-tags. Moreover, Fabs can be readily attached to a PG-Fc dimer module which acts as the core unit to produce plug-and-play IgG-like assemblies, and the utility can be further expanded to produce bispecific analogs using the "knobs into holes" strategy. These core PG-Fc dimer modules can be made and stored in bulk to produce off-the-shelf customized IgG entities in minutes, not days or weeks by just adding a Fab with the desired antigen specificity. In another application, the bispecific modalities form the building block for fabricating potent bispecific T-cell engagers (BiTEs), demonstrating their efficacy in cancer cell-killing assays. Additionally, the system can be adapted to include commercial antibodies as building blocks, greatly increasing the target space. Crystal structure analysis reveals that a few strategically positioned interactions engender the specificity between the Fab-PG variant pairs, requiring minimal changes to match the scaffolds for different possible combinations. This plug-and-play platform offers a user-friendly and versatile approach to enhance the functionality of antibody-based reagents in cell biology research.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Ngo, Wayne; Peukes, Julia; Baldwin, Alisha; Xue, Zhiwei Wayne; Hwang, Sidney; Stickels, Robert R; Lin, Zhi; Satpathy, Ansuman T; Wells, James A; Schekman, Randy; Nogales, Eva; Doudna, Jennifer A
Mechanism-guided engineering of a minimal biological particle for genome editing Journal Article
In: Proc Natl Acad Sci U S A, vol. 122, no. 1, pp. e2413519121, 2025, ISSN: 1091-6490.
@article{pmid39793042,
title = {Mechanism-guided engineering of a minimal biological particle for genome editing},
author = {Wayne Ngo and Julia Peukes and Alisha Baldwin and Zhiwei Wayne Xue and Sidney Hwang and Robert R Stickels and Zhi Lin and Ansuman T Satpathy and James A Wells and Randy Schekman and Eva Nogales and Jennifer A Doudna},
doi = {10.1073/pnas.2413519121},
issn = {1091-6490},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Proc Natl Acad Sci U S A},
volume = {122},
number = {1},
pages = {e2413519121},
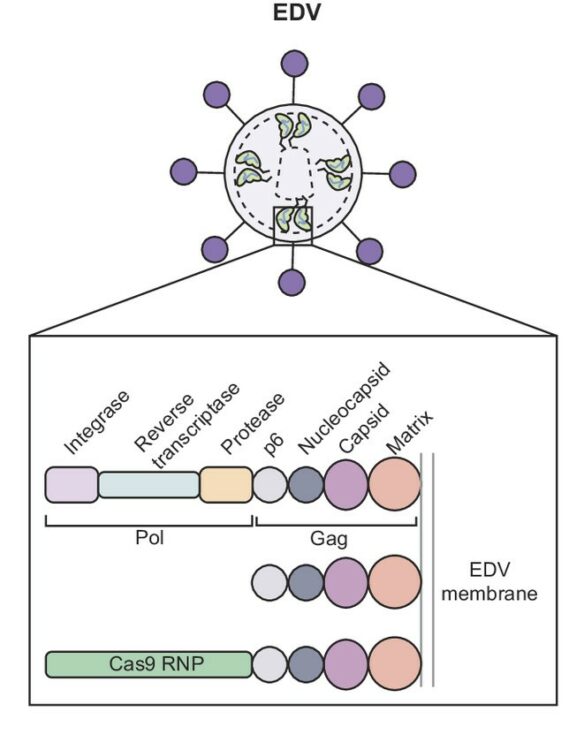
abstract = {The widespread application of genome editing to treat and cure disease requires the delivery of genome editors into the nucleus of target cells. Enveloped delivery vehicles (EDVs) are engineered virally derived particles capable of packaging and delivering CRISPR-Cas9 ribonucleoproteins (RNPs). However, the presence of lentiviral genome encapsulation and replication proteins in EDVs has obscured the underlying delivery mechanism and precluded particle optimization. Here, we show that Cas9 RNP nuclear delivery is independent of the native lentiviral capsid structure. Instead, EDV-mediated genome editing activity corresponds directly to the number of nuclear localization sequences on the Cas9 enzyme. EDV structural analysis using cryo-electron tomography and small molecule inhibitors guided the removal of ~80% of viral residues, creating a minimal EDV (miniEDV) that retains full RNP delivery capability. MiniEDVs are 25% smaller yet package equivalent amounts of Cas9 RNPs relative to the original EDVs and demonstrated increased editing in cell lines and therapeutically relevant primary human T cells. These results show that virally derived particles can be streamlined to create efficacious genome editing delivery vehicles with simpler production and manufacturing.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Górniak, Ireneusz; Stephens, Zachery; Erramilli, Satchal K; Gawda, Tomasz; Kossiakoff, Anthony A; Zimmer, Jochen
Structural insights into translocation and tailored synthesis of hyaluronan Journal Article
In: Nat Struct Mol Biol, vol. 32, no. 1, pp. 161–171, 2025, ISSN: 1545-9985.
@article{pmid39322765,
title = {Structural insights into translocation and tailored synthesis of hyaluronan},
author = {Ireneusz Górniak and Zachery Stephens and Satchal K Erramilli and Tomasz Gawda and Anthony A Kossiakoff and Jochen Zimmer},
doi = {10.1038/s41594-024-01389-1},
issn = {1545-9985},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Nat Struct Mol Biol},
volume = {32},
number = {1},
pages = {161--171},
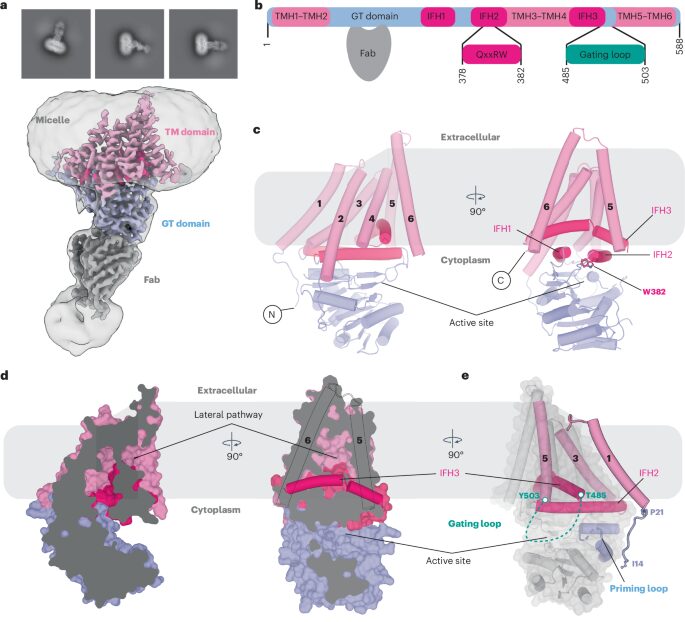
abstract = {Hyaluronan (HA) is an essential component of the vertebrate extracellular matrix. It is a heteropolysaccharide of N-acetylglucosamine (GlcNAc) and glucuronic acid (GlcA) reaching several megadaltons in healthy tissues. HA is synthesized and translocated in a coupled reaction by HA synthase (HAS). Here, structural snapshots of HAS provide insights into HA biosynthesis, from substrate recognition to HA elongation and translocation. We monitor the extension of a GlcNAc primer with GlcA, reveal the coordination of the uridine diphosphate product by a conserved gating loop and capture the opening of a translocation channel to coordinate a translocating HA polymer. Furthermore, we identify channel-lining residues that modulate HA product lengths. Integrating structural and biochemical analyses suggests an avenue for polysaccharide engineering based on finely tuned enzymatic activity and HA coordination.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
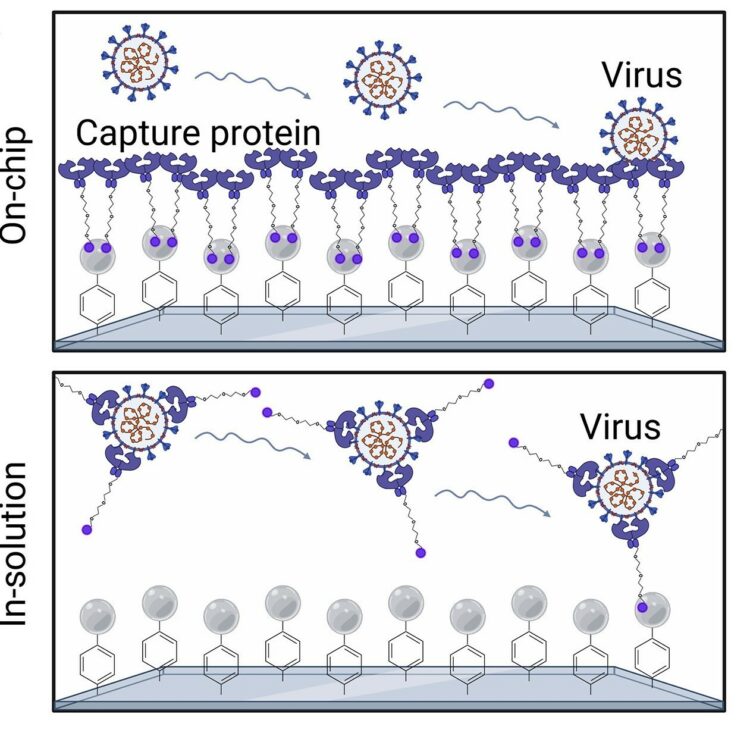
Rabe, Daniel C; Choudhury, Adarsh; Lee, Dasol; Luciani, Evelyn G; Ho, Uyen K; Clark, Alex E; Glasgow, Jeffrey E; Veiga, Sara; Michaud, William A; Capen, Diane; Flynn, Elizabeth A; Hartmann, Nicola; Garretson, Aaron F; Muzikansky, Alona; Goldberg, Marcia B; Kwon, Douglas S; Yu, Xu; Carlin, Aaron F; Theriault, Yves; Wells, James A; Lennerz, Jochen K; Lai, Peggy S; Rabi, Sayed Ali; Hoang, Anh N; Boland, Genevieve M; Stott, Shannon L
Ultrasensitive detection of intact SARS-CoV-2 particles in complex biofluids using microfluidic affinity capture Journal Article
In: Sci Adv, vol. 11, no. 2, pp. eadh1167, 2025, ISSN: 2375-2548.
@article{pmid39792670,
title = {Ultrasensitive detection of intact SARS-CoV-2 particles in complex biofluids using microfluidic affinity capture},
author = {Daniel C Rabe and Adarsh Choudhury and Dasol Lee and Evelyn G Luciani and Uyen K Ho and Alex E Clark and Jeffrey E Glasgow and Sara Veiga and William A Michaud and Diane Capen and Elizabeth A Flynn and Nicola Hartmann and Aaron F Garretson and Alona Muzikansky and Marcia B Goldberg and Douglas S Kwon and Xu Yu and Aaron F Carlin and Yves Theriault and James A Wells and Jochen K Lennerz and Peggy S Lai and Sayed Ali Rabi and Anh N Hoang and Genevieve M Boland and Shannon L Stott},
doi = {10.1126/sciadv.adh1167},
issn = {2375-2548},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Sci Adv},
volume = {11},
number = {2},
pages = {eadh1167},
abstract = {Measuring virus in biofluids is complicated by confounding biomolecules coisolated with viral nucleic acids. To address this, we developed an affinity-based microfluidic device for specific capture of intact severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Our approach used an engineered angiotensin-converting enzyme 2 to capture intact virus from plasma and other complex biofluids. Our device leverages a staggered herringbone pattern, nanoparticle surface coating, and processing conditions to achieve detection of as few as 3 viral copies per milliliter. We further validated our microfluidic assay on 103 plasma, 36 saliva, and 29 stool samples collected from unique patients with COVID-19, showing SARS-CoV-2 detection in 72% of plasma samples. Longitudinal monitoring in the plasma revealed our device's capacity for ultrasensitive detection of active viral infections over time. Our technology can be adapted to target other viruses using relevant cell entry molecules for affinity capture. This versatility underscores the potential for widespread application in viral load monitoring and disease management.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}