Publications: 2024
2024
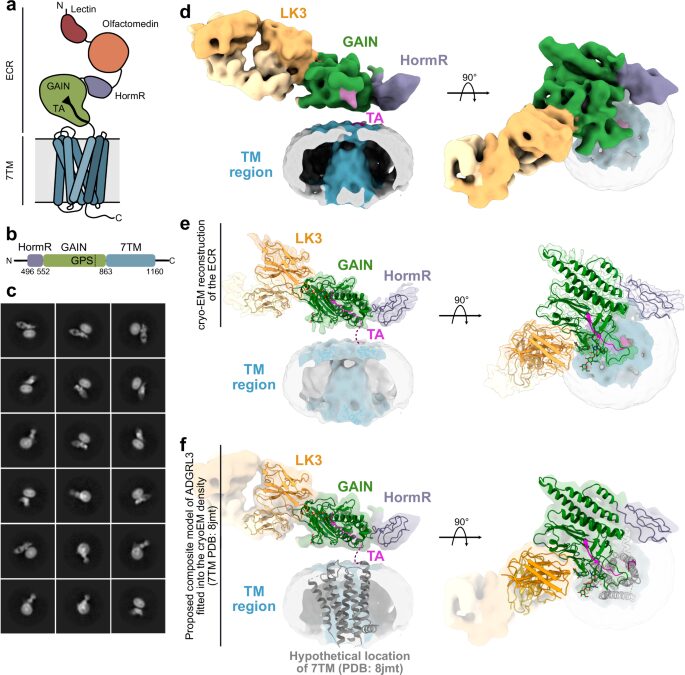
Kordon, Szymon P; Cechova, Kristina; Bandekar, Sumit J; Leon, Katherine; Dutka, Przemysław; Siffer, Gracie; Kossiakoff, Anthony A; Vafabakhsh, Reza; Araç, Demet
Conformational coupling between extracellular and transmembrane domains modulates holo-adhesion GPCR function Journal Article
In: Nat Commun, vol. 15, no. 1, pp. 10545, 2024, ISSN: 2041-1723.
@article{pmid39627215,
title = {Conformational coupling between extracellular and transmembrane domains modulates holo-adhesion GPCR function},
author = {Szymon P Kordon and Kristina Cechova and Sumit J Bandekar and Katherine Leon and Przemysław Dutka and Gracie Siffer and Anthony A Kossiakoff and Reza Vafabakhsh and Demet Araç},
doi = {10.1038/s41467-024-54836-4},
issn = {2041-1723},
year = {2024},
date = {2024-12-01},
urldate = {2024-12-01},
journal = {Nat Commun},
volume = {15},
number = {1},
pages = {10545},
abstract = {Adhesion G Protein-Coupled Receptors (aGPCRs) are key cell-adhesion molecules involved in numerous physiological functions. aGPCRs have large multi-domain extracellular regions (ECRs) containing a conserved GAIN domain that precedes their seven-pass transmembrane domain (7TM). Ligand binding and mechanical force applied on the ECR regulate receptor function. However, how the ECR communicates with the 7TM remains elusive, because the relative orientation and dynamics of the ECR and 7TM within a holoreceptor is unclear. Here, we describe the cryo-EM reconstruction of an aGPCR, Latrophilin3/ADGRL3, and reveal that the GAIN domain adopts a parallel orientation to the transmembrane region and has constrained movement. Single-molecule FRET experiments unveil three slow-exchanging FRET states of the ECR relative to the transmembrane region within the holoreceptor. GAIN-targeted antibodies, and cancer-associated mutations at the GAIN-7TM interface, alter FRET states, cryo-EM conformations, and receptor signaling. Altogether, this data demonstrates conformational and functional coupling between the ECR and 7TM, suggesting an ECR-mediated mechanism for aGPCR activation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
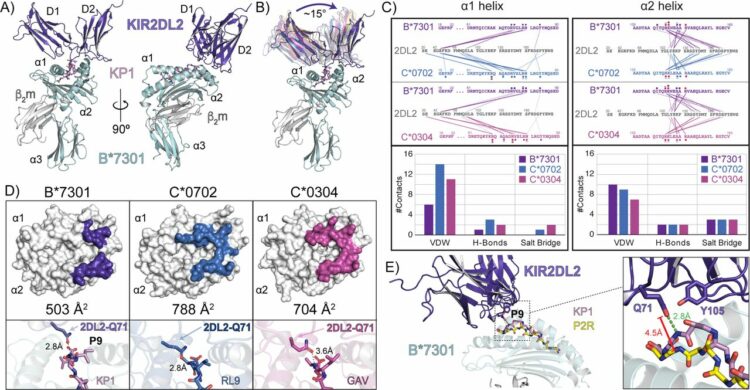
Ross, Philipp; Hilton, Hugo G; Lodwick, Jane; Slezak, Tomasz; Guethlein, Lisbeth A; McMurtrey, Curtis P; Han, Alex S; Nielsen, Morten; Yong, Daniel; Dulberger, Charles L; Nolan, Kristof T; Roy, Sobhan; Castro, Caitlin D; Hildebrand, William H; Zhao, Minglei; Kossiakoff, Anthony; Parham, Peter; Adams, Erin J
Molecular characterization of the archaic HLA-B*73:01 allele reveals presentation of a unique peptidome and skewed engagement by KIR2DL2 Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39651149,
title = {Molecular characterization of the archaic HLA-B*73:01 allele reveals presentation of a unique peptidome and skewed engagement by KIR2DL2},
author = {Philipp Ross and Hugo G Hilton and Jane Lodwick and Tomasz Slezak and Lisbeth A Guethlein and Curtis P McMurtrey and Alex S Han and Morten Nielsen and Daniel Yong and Charles L Dulberger and Kristof T Nolan and Sobhan Roy and Caitlin D Castro and William H Hildebrand and Minglei Zhao and Anthony Kossiakoff and Peter Parham and Erin J Adams},
doi = {10.1101/2024.11.25.625330},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {HLA class I alleles of archaic origin may have been retained in modern humans because they provide immunity against diseases to which archaic humans had evolved resistance. According to this model, archaic introgressed alleles were somehow distinct from those that evolved in African populations. Here we show that HLA-B*73:01, a rare allotype with putative archaic origins, has a relatively rare peptide binding motif with an unusually long-tailed peptide length distribution. We also find that HLA-B*73:01 combines a restricted and unique peptidome with high-cell surface expression, characteristics that make it well-suited to combat one or a number of closely-related pathogens. Furthermore, a crystal structure of HLA-B*73:01 in complex with KIR2DL2 highlights differences from previously solved structures with HLA-C molecules. These molecular characteristics distinguish HLA-B*73:01 from other HLA class I alleles previously investigated and may have provided early modern human migrants that inherited this allele with a selective advantage as they colonized Europe and Asia.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
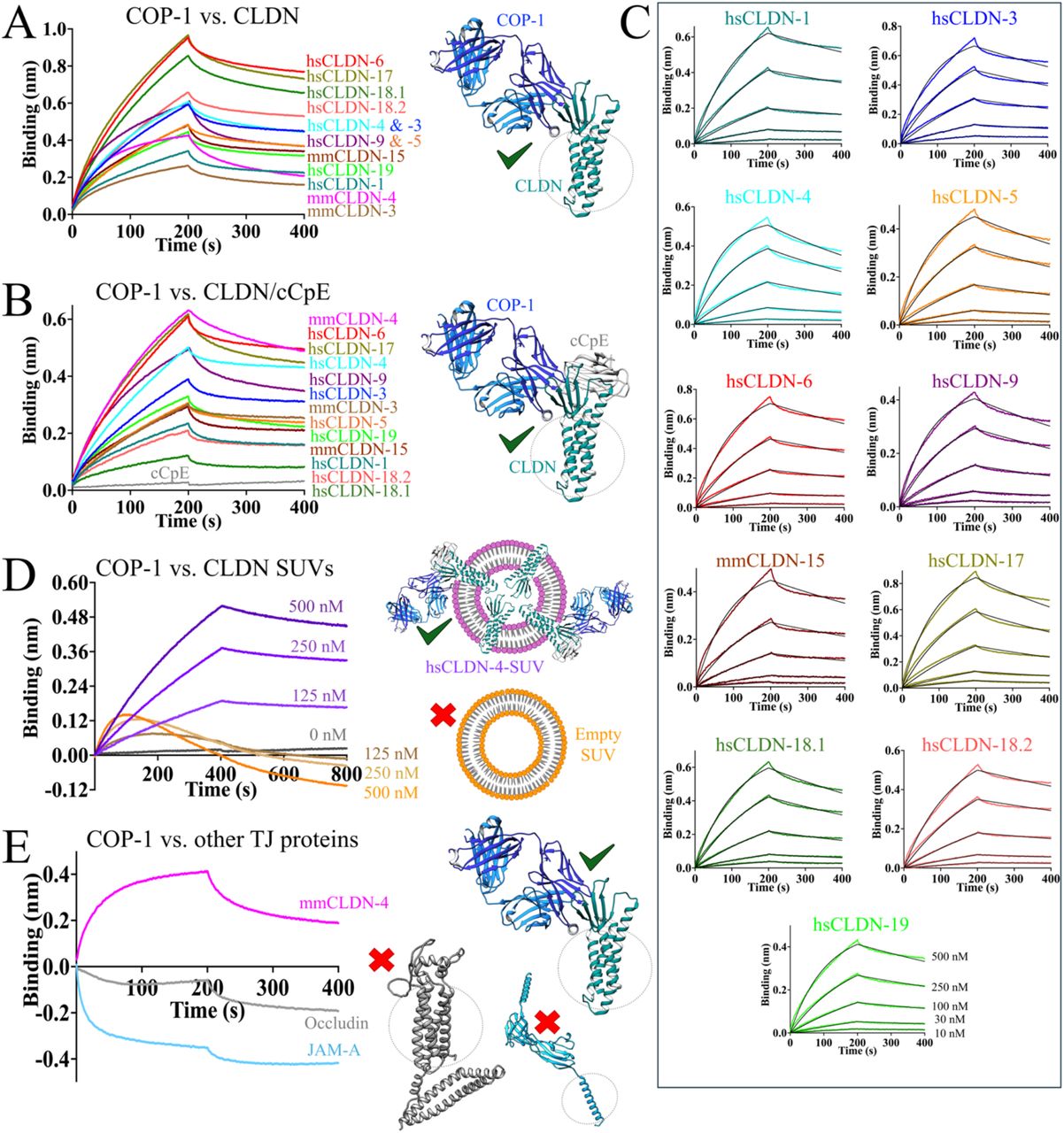
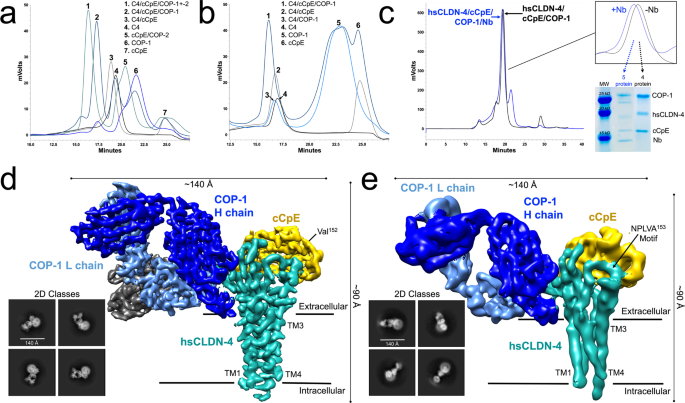
Ogbu, Chinemerem P; Mandriota, Alexandria M; Liu, Xiangdong; de Las Alas, Mason; Kapoor, Srajan; Choudhury, Jagrity; Kossiakoff, Anthony A; Duffey, Michael E; Vecchio, Alex J
Biophysical Basis of Paracellular Barrier Modulation by a Pan-Claudin-Binding Molecule Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39605593,
title = {Biophysical Basis of Paracellular Barrier Modulation by a Pan-Claudin-Binding Molecule},
author = {Chinemerem P Ogbu and Alexandria M Mandriota and Xiangdong Liu and Mason de Las Alas and Srajan Kapoor and Jagrity Choudhury and Anthony A Kossiakoff and Michael E Duffey and Alex J Vecchio},
doi = {10.1101/2024.11.10.622873},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {Claudins are a 27-member protein family that form and fortify specialized cell contacts in endothelium and epithelium called tight junctions. Tight junctions restrict paracellular transport across tissues by forming molecular barriers between cells. Claudin-binding molecules thus hold promise for modulating tight junction permeability to deliver drugs or as therapeutics to treat tight junction-linked disease. The development of claudin-binding molecules, however, is hindered by their intractability and small targetable surfaces. Here, we determine that a synthetic antibody fragment (sFab) we developed binds directly to 10 claudin subtypes with nanomolar affinity by targeting claudin's paracellular-exposed surface. Application of this sFab to cells that model intestinal epithelium show that it opens the paracellular barrier comparable to a known, but application limited, tight junction modulator. This novel pan-claudin-binding molecule can probe claudin or tight junction structure and holds potential as a broad modulator of tight junction permeability for basic or translational applications.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
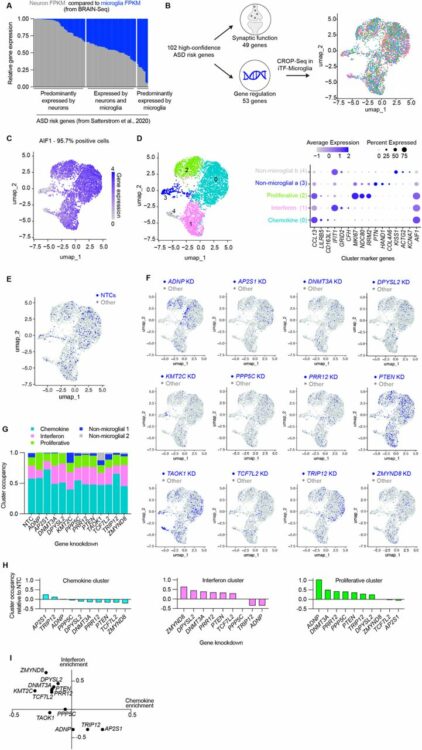
Teter, Olivia M; McQuade, Amanda; Hagan, Venus; Liang, Weiwei; Dräger, Nina M; Sattler, Sydney M; Holmes, Brandon B; Castillo, Vincent Cele; Papakis, Vasileios; Leng, Kun; Boggess, Steven; Nowakowski, Tomasz J; Wells, James; Kampmann, Martin
CRISPRi-based screen of Autism Spectrum Disorder risk genes in microglia uncovers roles of in microglia endocytosis and synaptic pruning Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39605704,
title = {CRISPRi-based screen of Autism Spectrum Disorder risk genes in microglia uncovers roles of in microglia endocytosis and synaptic pruning},
author = {Olivia M Teter and Amanda McQuade and Venus Hagan and Weiwei Liang and Nina M Dräger and Sydney M Sattler and Brandon B Holmes and Vincent Cele Castillo and Vasileios Papakis and Kun Leng and Steven Boggess and Tomasz J Nowakowski and James Wells and Martin Kampmann},
doi = {10.1101/2024.06.01.596962},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {Autism Spectrum Disorders (ASD) are a set of neurodevelopmental disorders with complex biology. The identification of ASD risk genes from exome-wide association studies and de novo variation analyses has enabled mechanistic investigations into how ASD-risk genes alter development. Most functional genomics studies have focused on the role of these genes in neurons and neural progenitor cells. However, roles for ASD risk genes in other cell types are largely uncharacterized. There is evidence from postmortem tissue that microglia, the resident immune cells of the brain, appear activated in ASD. Here, we used CRISPRi-based functional genomics to systematically assess the impact of ASD risk gene knockdown on microglia activation and phagocytosis. We developed an iPSC-derived microglia-neuron coculture system and high-throughput flow cytometry readout for synaptic pruning to enable parallel CRISPRi-based screening of phagocytosis of beads, synaptosomes, and synaptic pruning. Our screen identified , a high-confidence ASD risk genes, as a modifier of microglial synaptic pruning. We found that microglia with ADNP loss have altered endocytic trafficking, remodeled proteomes, and increased motility in coculture.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Martyn, Gregory D; Kalagiri, Rajasree; Veggiani, Gianluca; Stanfield, Robyn L; Choudhuri, Indrani; Sala, Margaux; Meisenhelder, Jill; Chen, Chao; Biswas, Avik; Levy, Ronald M; Lyumkis, Dmitry; Wilson, Ian A; Hunter, Tony; Sidhu, Sachdev S
Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39574610,
title = {Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody},
author = {Gregory D Martyn and Rajasree Kalagiri and Gianluca Veggiani and Robyn L Stanfield and Indrani Choudhuri and Margaux Sala and Jill Meisenhelder and Chao Chen and Avik Biswas and Ronald M Levy and Dmitry Lyumkis and Ian A Wilson and Tony Hunter and Sachdev S Sidhu},
doi = {10.1101/2024.11.04.621849},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {Histidine phosphorylation (pHis) is a non-canonical post-translational modification (PTM) that is historically understudied due to a lack of robust reagents that are required for its investigation, such as high affinity pHis-specific antibodies. Engineering pHis-specific antibodies is very challenging due to the labile nature of the phosphoramidate (P-N) bond and the stringent requirements for selective recognition of the two isoforms, 1-phosphohistidine (1-pHis) and 3-phosphohistidine (3-pHis). Here, we present a strategy for engineering of antibodies for detection of native 3-pHis targets. Specifically, we humanized the rabbit SC44-8 anti-3-pTza (a stable 3-pHis mimetic) mAb into a scaffold (herein referred to as hSC44) that was suitable for phage display. We then constructed six unique Fab phage-displayed libraries using the hSC44 scaffold and selected high affinity 3-pHis binders. Our selection strategy was carefully designed to enrich antibodies that bound 3-pHis with high affinity and had specificity for 3-pHis versus 3-pTza. hSC44.20N32F, the best engineered antibody, has an ~10-fold higher affinity for 3-pHis than the parental hSC44. Eleven new Fab structures, including the first reported antibody-pHis peptide structures were solved by X-ray crystallography. Structural and quantum mechanical calculations provided molecular insights into 3-pHis and 3-pTza discrimination by different hSC44 variants and their affinity increase obtained through engineering. Furthermore, we demonstrate the utility of these newly developed high-affinity 3-pHis-specific antibodies for recognition of pHis proteins in mammalian cells by immunoblotting and immunofluorescence staining. Overall, our work describes a general method for engineering PTM-specific antibodies and provides a set of novel antibodies for further investigations of the role of 3-pHis in cell biology.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Rathnayake, Sewwandi S; Erramilli, Satchal K; Kossiakoff, Anthony A; Vecchio, Alex J
Cryo-EM structures of Clostridium perfringens enterotoxin bound to its human receptor, claudin-4 Journal Article
In: Structure, vol. 32, no. 11, pp. 1936–1951.e5, 2024, ISSN: 1878-4186.
@article{pmid39383874,
title = {Cryo-EM structures of Clostridium perfringens enterotoxin bound to its human receptor, claudin-4},
author = {Sewwandi S Rathnayake and Satchal K Erramilli and Anthony A Kossiakoff and Alex J Vecchio},
doi = {10.1016/j.str.2024.09.015},
issn = {1878-4186},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {Structure},
volume = {32},
number = {11},
pages = {1936--1951.e5},
abstract = {Clostridium perfringens enterotoxin (CpE) causes prevalent and deadly gastrointestinal disorders. CpE binds to receptors called claudins on the apical surfaces of small intestinal epithelium. Claudins normally regulate paracellular transport but are hijacked from doing so by CpE and are instead led to form claudin/CpE complexes. Claudin/CpE complexes are the building blocks of oligomeric β-barrel pores that penetrate the plasma membrane and induce gut cytotoxicity. Here, we present the structures of CpE in complex with its native claudin receptor in humans, claudin-4, using cryogenic electron microscopy. The structures reveal the architecture of the claudin/CpE complex, the residues used in binding, the orientation of CpE relative to the membrane, and CpE-induced changes to claudin-4. Further, structures and modeling allude to the biophysical procession from claudin/CpE complexes to cytotoxic β-barrel pores during pathogenesis. In full, this work proposes a model of claudin/CpE assembly and provides strategies to obstruct its formation to treat CpE diseases.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
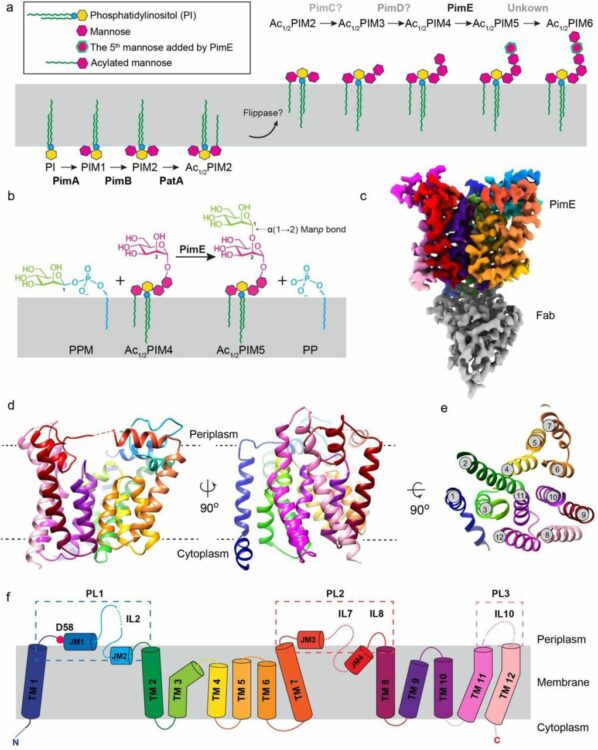
Liu, Yaqi; Brown, Chelsea M; Borges, Nuno; Nobre, Rodrigo N; Erramilli, Satchal; Dufrisne, Meagan Belcher; Kloss, Brian; Giacometti, Sabrina; Esteves, Ana M; Timoteo, Cristina G; Tokarz, Piotr; Cater, Rosemary; Morita, Yasu S; Kossiakoff, Anthony A; Santos, Helena; Stansfeld, Phillip J; Nygaard, Rie; Mancia, Filippo
Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39345506,
title = {Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE},
author = {Yaqi Liu and Chelsea M Brown and Nuno Borges and Rodrigo N Nobre and Satchal Erramilli and Meagan Belcher Dufrisne and Brian Kloss and Sabrina Giacometti and Ana M Esteves and Cristina G Timoteo and Piotr Tokarz and Rosemary Cater and Yasu S Morita and Anthony A Kossiakoff and Helena Santos and Phillip J Stansfeld and Rie Nygaard and Filippo Mancia},
doi = {10.1101/2024.09.17.613550},
issn = {2692-8205},
year = {2024},
date = {2024-09-01},
urldate = {2024-09-01},
journal = {bioRxiv},
abstract = {Tuberculosis (TB), exceeded in mortality only by COVID-19 among global infectious diseases, is caused by Mycobacterium tuberculosis (Mtb). The pathogenicity of Mtb is largely attributed to its complex cell envelope, which includes a class of glycolipids called phosphatidyl-myo-inositol mannosides (PIMs), found uniquely in mycobacteria and its related corynebacterineae. These glycolipids maintain the integrity of the mycobacterial cell envelope, regulate its permeability, and mediate host-pathogen interactions. PIMs consist of a phosphatidyl-myo-inositol core decorated with one to six mannose residues and up to four acyl chains. The mannosyltransferase PimE catalyzes the transfer of the fifth PIM mannose residue from a polyprenyl phosphate-mannose (PPM) donor. This step in the biosynthesis of higher-order PIMs contributes to the proper assembly and function of the mycobacterial cell envelope; however, the structural basis for substrate recognition and the catalytic mechanism of PimE remain poorly understood. Here, we present the cryo-electron microscopy (cryo-EM) structures of PimE from Mycobacterium abscessus captured in its apo form and in a product-bound complex with the reaction product Ac1PIM5 and the by-product polyprenyl phosphate (PP), determined at 3.0 Å and 3.5 Å, respectively. The structures reveal the active site within a distinctive binding cavity that accommodates both donor and acceptor substrates/products. Within the cavity, we identified residues involved in substrate coordination and catalysis, which we confirmed through in vitro enzymatic assays and further validated by in vivo complementation experiments. Molecular dynamics simulations were applied to identify the access pathways and the dynamics involved in substrate binding. Integrating structural, biochemical, genetic, and computational experiments, our study provides comprehensive insights into how PimE functions, opening potential avenues for development of novel anti-TB therapeutics.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
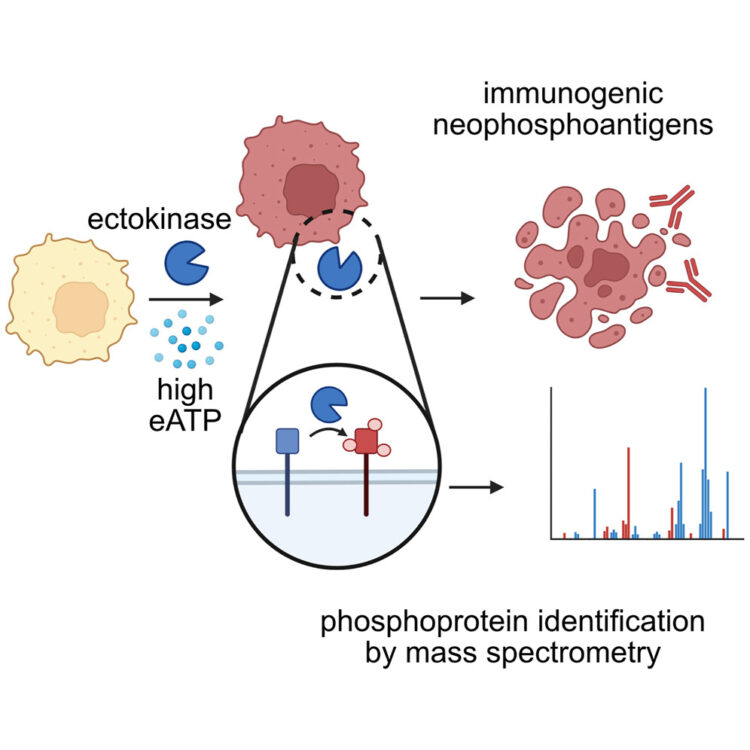
Delaveris, Corleone S; Kong, Sophie; Glasgow, Jeff; Loudermilk, Rita P; Kirkemo, Lisa L; Zhao, Fangzhu; Salangsang, Fernando; Phojanakong, Paul; Serrano, Juan Antonio Camara; Steri, Veronica; Wells, James A
Chemoproteomics reveals immunogenic and tumor-associated cell surface substrates of ectokinase CK2α Journal Article
In: Cell Chem Biol, vol. 31, no. 9, pp. 1729–1739.e9, 2024, ISSN: 2451-9448.
@article{pmid39178841,
title = {Chemoproteomics reveals immunogenic and tumor-associated cell surface substrates of ectokinase CK2α},
author = {Corleone S Delaveris and Sophie Kong and Jeff Glasgow and Rita P Loudermilk and Lisa L Kirkemo and Fangzhu Zhao and Fernando Salangsang and Paul Phojanakong and Juan Antonio Camara Serrano and Veronica Steri and James A Wells},
doi = {10.1016/j.chembiol.2024.07.018},
issn = {2451-9448},
year = {2024},
date = {2024-09-01},
urldate = {2024-09-01},
journal = {Cell Chem Biol},
volume = {31},
number = {9},
pages = {1729--1739.e9},
abstract = {Foreign epitopes for immune recognition provide the basis of anticancer immunity. Due to the high concentration of extracellular adenosine triphosphate in the tumor microenvironment, we hypothesized that extracellular kinases (ectokinases) could have dysregulated activity and introduce aberrant phosphorylation sites on cell surface proteins. We engineered a cell-tethered version of the extracellular kinase CK2α, demonstrated it was active on cells under tumor-relevant conditions, and profiled its substrate scope using a chemoproteomic workflow. We then demonstrated that mice developed polyreactive antisera in response to syngeneic tumor cells that had been subjected to surface hyperphosphorylation with CK2α. Interestingly, these mice developed B cell and CD4 T cell responses in response to these antigens but failed to develop a CD8 T cell response. This work provides a workflow for probing the extracellular phosphoproteome and demonstrates that extracellular phosphoproteins are immunogenic even in a syngeneic system.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Samassekou, Kadidia; Garland-Kuntz, Elisabeth E; Ohri, Vaani; Fisher, Isaac J; Erramilli, Satchal K; Muralidharan, Kaushik; Bogdan, Livia M; Gick, Abigail M; Kossiakoff, Anthony A; Lyon, Angeline M
Cryo-EM Structure of Phospholipase Cε Defines N-terminal Domains and their Roles in Activity Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39314324,
title = {Cryo-EM Structure of Phospholipase Cε Defines N-terminal Domains and their Roles in Activity},
author = {Kadidia Samassekou and Elisabeth E Garland-Kuntz and Vaani Ohri and Isaac J Fisher and Satchal K Erramilli and Kaushik Muralidharan and Livia M Bogdan and Abigail M Gick and Anthony A Kossiakoff and Angeline M Lyon},
doi = {10.1101/2024.09.11.612521},
issn = {2692-8205},
year = {2024},
date = {2024-09-01},
urldate = {2024-09-01},
journal = {bioRxiv},
abstract = {Phospholipase Cε (PLCε) increases intracellular Ca and protein kinase C (PKC) activity in the cardiovascular system in response to stimulation of G protein coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs). The ability of PLCε to respond to these diverse inputs is due, in part, to multiple, conformationally dynamic regulatory domains. However, this heterogeneity has also limited structural studies of the lipase to either individual domains or its catalytic core. Here, we report the 3.9 Å reconstruction of the largest fragment of PLCε to date in complex with an antigen binding fragment (Fab). The structure reveals that PLCε contains a pleckstrin homology (PH) domain and four tandem EF hands, including subfamily-specific insertions and intramolecular interactions with the catalytic core. The structure, together with a model of the holoenzyme, suggest that part of the N-terminus and PH domain form a continuous surface that could engage cytoplasmic leaflets of the plasma and perinuclear membranes, contributing to activity. Functional characterization of this surface confirm it is critical for maximum basal and G protein-stimulated activities. This study provides new insights into the autoinhibited, basal conformation of PLCε and the first mechanistic insights into how it engages cellular membranes for activity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
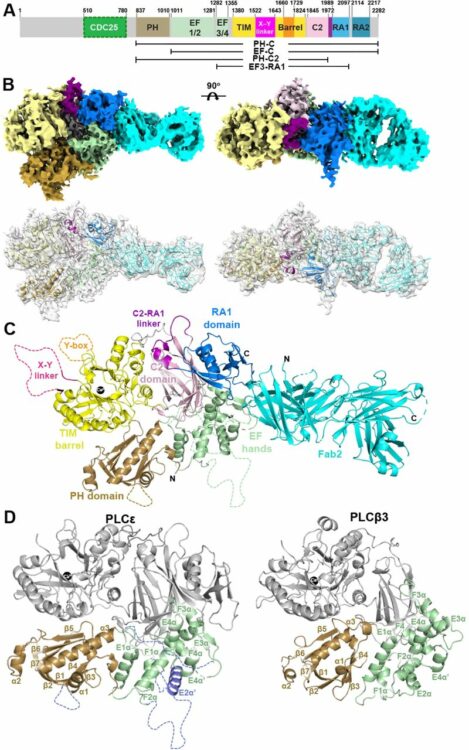
Liu, Yaqi; Brown, Chelsea M; Erramilli, Satchal; Su, Yi-Chia; Tseng, Po-Sen; Wang, Yu-Jen; Duong, Nam Ha; Tokarz, Piotr; Kloss, Brian; Han, Cheng-Ruei; Chen, Hung-Yu; Rodrigues, Jose; Archer, Margarida; Lowary, Todd L; Kossiakoff, Anthony A; Stansfeld, Phillip J; Nygaard, Rie; Mancia, Filippo
Structural insights into terminal arabinosylation biosynthesis of the mycobacterial cell wall arabinan Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39345558,
title = {Structural insights into terminal arabinosylation biosynthesis of the mycobacterial cell wall arabinan},
author = {Yaqi Liu and Chelsea M Brown and Satchal Erramilli and Yi-Chia Su and Po-Sen Tseng and Yu-Jen Wang and Nam Ha Duong and Piotr Tokarz and Brian Kloss and Cheng-Ruei Han and Hung-Yu Chen and Jose Rodrigues and Margarida Archer and Todd L Lowary and Anthony A Kossiakoff and Phillip J Stansfeld and Rie Nygaard and Filippo Mancia},
doi = {10.1101/2024.09.17.613533},
issn = {2692-8205},
year = {2024},
date = {2024-09-01},
urldate = {2024-09-01},
journal = {bioRxiv},
abstract = {The emergence of drug-resistant strains exacerbates the global challenge of tuberculosis caused by Mycobacterium tuberculosis (Mtb). Central to the pathogenicity of Mtb is its complex cell envelope, which serves as a barrier against both immune system and pharmacological attacks. Two key components of this envelope, arabinogalactan (AG) and lipoarabinomannan (LAM) are complex polysaccharides that contain integral arabinan domains important for cell wall structural and functional integrity. The arabinofuranosyltransferase AftB terminates the synthesis of these arabinan domains by catalyzing the addition of the addition of β-(1→2)-linked terminal arabinofuranose residues. Here, we present the cryo-EM structures of Mycobacterium chubuense AftB in its apo and donor substrate analog-bound form, determined to 2.9 Å and 3.4 Å resolution, respectively. Our structures reveal that AftB has a GT-C fold transmembrane (TM) domain comprised of eleven TM helices and a periplasmic cap domain. AftB has an irregular tube-shaped cavity that bridges the two proposed substrate binding sites. By integrating structural analysis, biochemical assays, and molecular dynamics simulations, we elucidate the molecular basis of the reaction mechanism of AftB and propose a model for catalysis.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
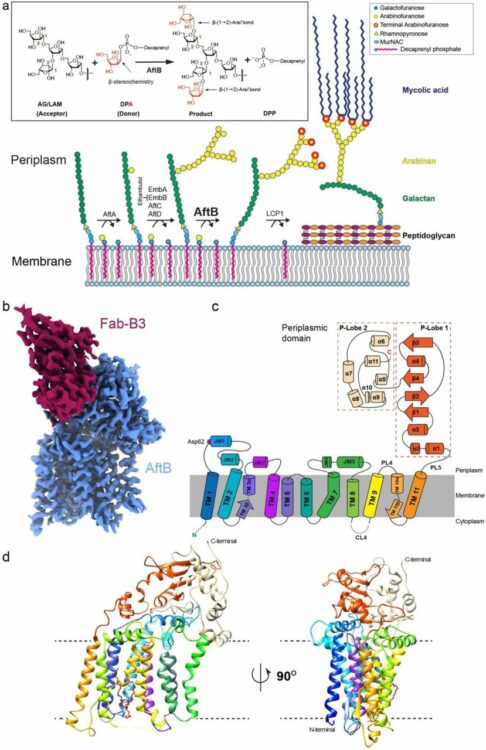
Knejski, Paweł P; Erramilli, Satchal K; Kossiakoff, Anthony A
Chaperone-assisted cryo-EM structure of PhuR reveals molecular basis for heme binding Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid37577460,
title = {Chaperone-assisted cryo-EM structure of PhuR reveals molecular basis for heme binding},
author = {Paweł P Knejski and Satchal K Erramilli and Anthony A Kossiakoff},
doi = {10.1101/2023.08.01.551527},
issn = {2692-8205},
year = {2024},
date = {2024-08-01},
urldate = {2024-08-01},
journal = {bioRxiv},
abstract = {Pathogenic bacteria, such as , depend on scavenging heme for the acquisition of iron, an essential nutrient. The TonB-dependent transporter (TBDT) PhuR is the major heme uptake protein in clinical isolates. However, a comprehensive understanding of heme recognition and TBDT transport mechanisms, especially PhuR, remains limited. In this study, we employed single-particle cryogenic electron microscopy (cryo-EM) and a phage display-generated synthetic antibody (sAB) as a fiducial marker to enable the determination of a high-resolution (2.5 Å) structure of PhuR with a bound heme. Notably, the structure reveals iron coordination by Y529 on a conserved extracellular loop, shedding light on the role of tyrosine in heme binding. Biochemical assays and negative-stain EM demonstrated that the sAB specifically targets the heme-bound state of PhuR. These findings provide insights into PhuR's heme binding and offer a template for developing conformation-specific sABs against outer membrane proteins (OMPs) for structure-function investigations.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
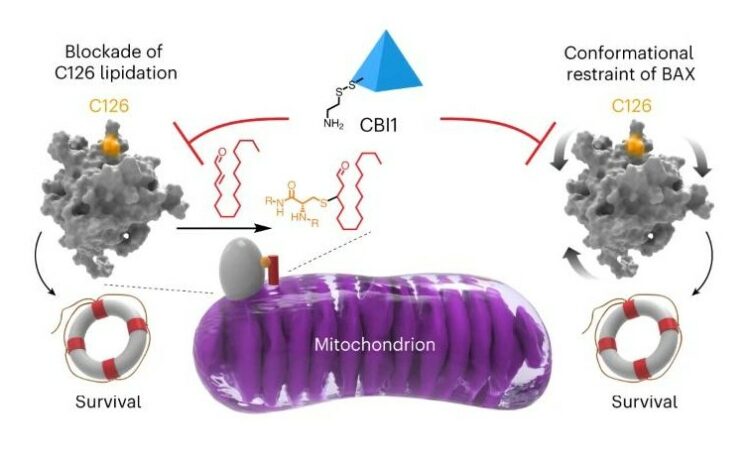
McHenry, Matthew W; Shi, Peiwen; Camara, Christina M; Cohen, Daniel T; Rettenmaier, T Justin; Adhikary, Utsarga; Gygi, Micah A; Yang, Ka; Gygi, Steven P; Wales, Thomas E; Engen, John R; Wells, James A; Walensky, Loren D
Covalent inhibition of pro-apoptotic BAX Journal Article
In: Nat Chem Biol, vol. 20, no. 8, pp. 1022–1032, 2024, ISSN: 1552-4469.
@article{pmid38233584,
title = {Covalent inhibition of pro-apoptotic BAX},
author = {Matthew W McHenry and Peiwen Shi and Christina M Camara and Daniel T Cohen and T Justin Rettenmaier and Utsarga Adhikary and Micah A Gygi and Ka Yang and Steven P Gygi and Thomas E Wales and John R Engen and James A Wells and Loren D Walensky},
doi = {10.1038/s41589-023-01537-6},
issn = {1552-4469},
year = {2024},
date = {2024-08-01},
urldate = {2024-08-01},
journal = {Nat Chem Biol},
volume = {20},
number = {8},
pages = {1022--1032},
abstract = {BCL-2-associated X protein (BAX) is a promising therapeutic target for activating or restraining apoptosis in diseases of pathologic cell survival or cell death, respectively. In response to cellular stress, BAX transforms from a quiescent cytosolic monomer into a toxic oligomer that permeabilizes the mitochondria, releasing key apoptogenic factors. The mitochondrial lipid trans-2-hexadecenal (t-2-hex) sensitizes BAX activation by covalent derivatization of cysteine 126 (C126). In this study, we performed a disulfide tethering screen to discover C126-reactive molecules that modulate BAX activity. We identified covalent BAX inhibitor 1 (CBI1) as a compound that selectively derivatizes BAX at C126 and inhibits BAX activation by triggering ligands or point mutagenesis. Biochemical and structural analyses revealed that CBI1 can inhibit BAX by a dual mechanism of action: conformational constraint and competitive blockade of lipidation. These data inform a pharmacologic strategy for suppressing apoptosis in diseases of unwanted cell death by covalent targeting of BAX C126.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Slezak, Tomasz; O'Leary, Kelly M; Li, Jinyang; Rohaim, Ahmed; Davydova, Elena K; Kossiakoff, Anthony A
Engineered Protein-G variants for plug-and-play applications Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39211271,
title = {Engineered Protein-G variants for plug-and-play applications},
author = {Tomasz Slezak and Kelly M O'Leary and Jinyang Li and Ahmed Rohaim and Elena K Davydova and Anthony A Kossiakoff},
doi = {10.1101/2024.08.06.606809},
issn = {2692-8205},
year = {2024},
date = {2024-08-01},
urldate = {2024-08-01},
journal = {bioRxiv},
abstract = {We have developed a portfolio of antibody-based modules that can be prefabricated as standalone units and snapped together in plug-and-play fashion to create uniquely powerful multifunctional assemblies. The basic building blocks are derived from multiple pairs of native and modified Fab scaffolds and protein G (PG) variants engineered by phage display to introduce high pair-wise specificity. The variety of possible Fab-PG pairings provides a highly orthogonal system that can be exploited to perform challenging cell biology operations in a straightforward manner. The simplest manifestation allows multiplexed antigen detection using PG variants fused to fluorescently labeled SNAP-tags. Moreover, Fabs can be readily attached to a PG-Fc dimer module which acts as the core unit to produce plug-and-play IgG-like assemblies, and the utility can be further expanded to produce bispecific analogs using the "knobs into holes" strategy. These core PG-Fc dimer modules can be made and stored in bulk to produce off-the-shelf customized IgG entities in minutes, not days or weeks by just adding a Fab with the desired antigen specificity. In another application, the bispecific modalities form the building block for fabricating potent Bispecific T-cell Engagers (BiTEs), demonstrating their efficacy in cancer cell-killing assays. Additionally, the system can be adapted to include commercial antibodies as building blocks, greatly increasing the target space. Crystal structure analysis reveals that a few strategically positioned interactions engender the specificity between the Fab-PG variant pairs, requiring minimal changes to match the scaffolds for different possible combinations. This plug-and-play platform offers a user-friendly and versatile approach to enhance the functionality of antibody-based reagents in cell biology research.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
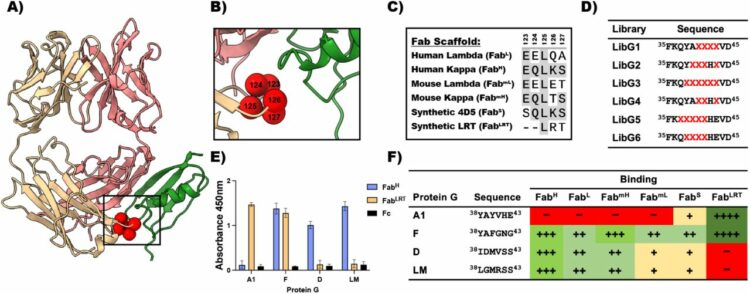
Bruce, Heather A.; Singer, Alexander U.; Blazer, Levi L.; Luu, Khanh; Ploder, Lynda; Pavlenco, Alevtina; Kurinov, Igor; Adams, Jarrett J.; Sidhu, Sachdev S.
In: Protein Science, vol. 33, no. 7, 2024, ISSN: 1469-896X.
@article{Bruce2024,
title = {Antigen‐binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones},
author = {Heather A. Bruce and Alexander U. Singer and Levi L. Blazer and Khanh Luu and Lynda Ploder and Alevtina Pavlenco and Igor Kurinov and Jarrett J. Adams and Sachdev S. Sidhu},
doi = {10.1002/pro.5081},
issn = {1469-896X},
year = {2024},
date = {2024-07-00},
urldate = {2024-07-00},
journal = {Protein Science},
volume = {33},
number = {7},
publisher = {Wiley},
abstract = {<jats:title>Abstract</jats:title><jats:p>It has been shown previously that a set of three modifications—termed S1, Crystal Kappa, and elbow—act synergistically to improve the crystallizability of an antigen‐binding fragment (Fab) framework. Here, we prepared a phage‐displayed library and performed crystallization screenings to identify additional substitutions—located near the heavy‐chain elbow region—which cooperate with the S1, Crystal Kappa, and elbow modifications to increase expression and improve crystallizability of the Fab framework even further. One substitution (K141Q) supports the signature Crystal Kappa‐mediated Fab:Fab crystal lattice packing interaction. Another substitution (E172G) improves the compatibility of the elbow modification with the Fab framework by alleviating some of the strain incurred by the shortened and bulkier elbow linker region. A third substitution (F170W) generates a split‐Fab conformation, resulting in a powerful crystal lattice packing interaction comprising the biological interaction interface between the variable heavy and light chain domains. In sum, we have used K141Q, E172G, and F170W substitutions—which complement the S1, Crystal Kappa, and elbow modifications—to generate a set of highly crystallizable Fab frameworks that can be used as chaperones to enable facile elucidation of Fab:antigen complex structures by x‐ray crystallography.</jats:p>},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
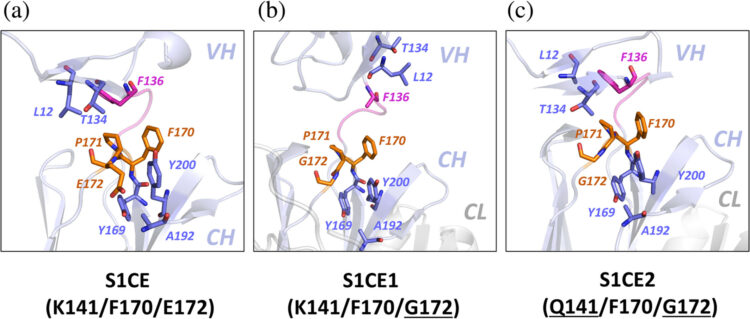
Pluvinage, John V.; Ngo, Thomas; Fouassier, Camille; McDonagh, Maura; Holmes, Brandon B.; Bartley, Christopher M.; Kondapavulur, Sravani; Hurabielle, Charlotte; Bodansky, Aaron; Pai, Vincent; Hinman, Sam; Aslanpour, Ava; Alvarenga, Bonny D.; Zorn, Kelsey C.; Zamecnik, Colin; McCann, Adrian; Asencor, Andoni I.; Huynh, Trung; Browne, Weston; Tubati, Asritha; Haney, Michael S.; Douglas, Vanja C.; Louine, Martineau; Cree, Bruce A. C.; Hauser, Stephen L.; Seeley, William; Baranzini, Sergio E.; Wells, James A.; Spudich, Serena; Farhadian, Shelli; Ramachandran, Prashanth S.; Gillum, Leslie; Hales, Chadwick M.; Zikherman, Julie; Anderson, Mark S.; Yazdany, Jinoos; Smith, Bryan; Nath, Avindra; Suh, Gina; Flanagan, Eoin P.; Green, Ari J.; Green, Ralph; Gelfand, Jeffrey M.; DeRisi, Joseph L.; Pleasure, Samuel J.; Wilson, Michael R.
Transcobalamin receptor antibodies in autoimmune vitamin B12 central deficiency Journal Article
In: Sci. Transl. Med., vol. 16, no. 753, 2024, ISSN: 1946-6242.
@article{Pluvinage2024,
title = {Transcobalamin receptor antibodies in autoimmune vitamin B12 central deficiency},
author = {John V. Pluvinage and Thomas Ngo and Camille Fouassier and Maura McDonagh and Brandon B. Holmes and Christopher M. Bartley and Sravani Kondapavulur and Charlotte Hurabielle and Aaron Bodansky and Vincent Pai and Sam Hinman and Ava Aslanpour and Bonny D. Alvarenga and Kelsey C. Zorn and Colin Zamecnik and Adrian McCann and Andoni I. Asencor and Trung Huynh and Weston Browne and Asritha Tubati and Michael S. Haney and Vanja C. Douglas and Martineau Louine and Bruce A.C. Cree and Stephen L. Hauser and William Seeley and Sergio E. Baranzini and James A. Wells and Serena Spudich and Shelli Farhadian and Prashanth S. Ramachandran and Leslie Gillum and Chadwick M. Hales and Julie Zikherman and Mark S. Anderson and Jinoos Yazdany and Bryan Smith and Avindra Nath and Gina Suh and Eoin P. Flanagan and Ari J. Green and Ralph Green and Jeffrey M. Gelfand and Joseph L. DeRisi and Samuel J. Pleasure and Michael R. Wilson},
doi = {10.1126/scitranslmed.adl3758},
issn = {1946-6242},
year = {2024},
date = {2024-06-26},
urldate = {2024-06-26},
journal = {Sci. Transl. Med.},
volume = {16},
number = {753},
publisher = {American Association for the Advancement of Science (AAAS)},
abstract = {Vitamin B12 is critical for hematopoiesis and myelination. Deficiency can cause neurologic deficits including loss of coordination and cognitive decline. However, diagnosis relies on measurement of vitamin B12 in the blood, which may not accurately reflect the concentration in the brain. Using programmable phage display, we identified an autoantibody targeting the transcobalamin receptor (CD320) in a patient with progressive tremor, ataxia, and scanning speech. Anti-CD320 impaired cellular uptake of cobalamin (B12) in vitro by depleting its target from the cell surface. Despite a normal serum concentration, B12 was nearly undetectable in her cerebrospinal fluid (CSF). Immunosuppressive treatment and high-dose systemic B12 supplementation were associated with increased B12 in the CSF and clinical improvement. Optofluidic screening enabled isolation of a patient-derived monoclonal antibody that impaired B12 transport across an in vitro model of the blood-brain barrier (BBB). Autoantibodies targeting the same epitope of CD320 were identified in seven other patients with neurologic deficits of unknown etiology, 6% of healthy controls, and 21.4% of a cohort of patients with neuropsychiatric lupus. In 132 paired serum and CSF samples, detection of anti-CD320 in the blood predicted B12 deficiency in the brain. However, these individuals did not display any hematologic signs of B12 deficiency despite systemic CD320 impairment. Using a genome-wide CRISPR screen, we found that the low-density lipoprotein receptor serves as an alternative B12 uptake pathway in hematopoietic cells. These findings dissect the tissue specificity of B12 transport and elucidate an autoimmune neurologic condition that may be amenable to immunomodulatory treatment and nutritional supplementation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Lan, Tong; Slezak, Tomasz; Pu, Jinyue; Zinkus-Boltz, Julia; Adhikari, Sarbani; Pekow, Joel R; Taneja, Vibha; Zuniga, Joaquin; Gómez-García, Itzel A; Regino-Zamarripa, Nora; Ahmed, Mushtaq; Khader, Shabaana A; Rubin, David T; Kossiakoff, Anthony A; Dickinson, Bryan C
Development of Luminescent Biosensors for Calprotectin Journal Article
In: ACS Chem Biol, vol. 19, no. 6, pp. 1250–1259, 2024, ISSN: 1554-8937.
@article{pmid38843544,
title = {Development of Luminescent Biosensors for Calprotectin},
author = {Tong Lan and Tomasz Slezak and Jinyue Pu and Julia Zinkus-Boltz and Sarbani Adhikari and Joel R Pekow and Vibha Taneja and Joaquin Zuniga and Itzel A Gómez-García and Nora Regino-Zamarripa and Mushtaq Ahmed and Shabaana A Khader and David T Rubin and Anthony A Kossiakoff and Bryan C Dickinson},
doi = {10.1021/acschembio.4c00265},
issn = {1554-8937},
year = {2024},
date = {2024-06-01},
urldate = {2024-06-01},
journal = {ACS Chem Biol},
volume = {19},
number = {6},
pages = {1250--1259},
abstract = {Calprotectin, a metal ion-binding protein complex, plays a crucial role in the innate immune system and has gained prominence as a biomarker for various intestinal and systemic inflammatory and infectious diseases, including inflammatory bowel disease (IBD) and tuberculosis (TB). Current clinical testing methods rely on enzyme-linked immunosorbent assays (ELISAs), limiting accessibility and convenience. In this study, we introduce the Fab-Enabled Split-luciferase Calprotectin Assay (FESCA), a novel quantitative method for calprotectin measurement. FESCA utilizes two new fragment antigen binding proteins (Fabs), CP16 and CP17, that bind to different epitopes of the calprotectin complex. These Fabs are fused with split NanoLuc luciferase fragments, enabling the reconstitution of active luciferase upon binding to calprotectin either in solution or in varied immobilized assay formats. FESCA's output luminescence can be measured with standard laboratory equipment as well as consumer-grade cell phone cameras. FESCA can detect physiologically relevant calprotectin levels across various sample types, including serum, plasma, and whole blood. Notably, FESCA can detect abnormally elevated native calprotectin from TB patients. In summary, FESCA presents a convenient, low-cost, and quantitative method for assessing calprotectin levels in various biological samples, with the potential to improve the diagnosis and monitoring of inflammatory diseases, especially in at-home or point-of-care settings.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Erramilli, Satchal K; Dominik, Pawel K; Ogbu, Chinemerem P; Kossiakoff, Anthony A; Vecchio, Alex J
Structural and biophysical insights into targeting of claudin-4 by a synthetic antibody fragment Journal Article
In: Commun Biol, vol. 7, no. 1, pp. 733, 2024, ISSN: 2399-3642.
@article{pmid38886509,
title = {Structural and biophysical insights into targeting of claudin-4 by a synthetic antibody fragment},
author = {Satchal K Erramilli and Pawel K Dominik and Chinemerem P Ogbu and Anthony A Kossiakoff and Alex J Vecchio},
doi = {10.1038/s42003-024-06437-6},
issn = {2399-3642},
year = {2024},
date = {2024-06-01},
urldate = {2024-06-01},
journal = {Commun Biol},
volume = {7},
number = {1},
pages = {733},
abstract = {Claudins are a 27-member family of ~25 kDa membrane proteins that integrate into tight junctions to form molecular barriers at the paracellular spaces between endothelial and epithelial cells. As the backbone of tight junction structure and function, claudins are attractive targets for modulating tissue permeability to deliver drugs or treat disease. However, structures of claudins are limited due to their small sizes and physicochemical properties-these traits also make therapy development a challenge. Here we report the development of a synthetic antibody fragment (sFab) that binds human claudin-4 and the determination of a high-resolution structure of it bound to claudin-4/enterotoxin complexes using cryogenic electron microscopy. Structural and biophysical results reveal this sFabs mechanism of select binding to human claudin-4 over other homologous claudins and establish the ability of sFabs to bind hard-to-target claudins to probe tight junction structure and function. The findings provide a framework for tight junction modulation by sFabs for tissue-selective therapies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
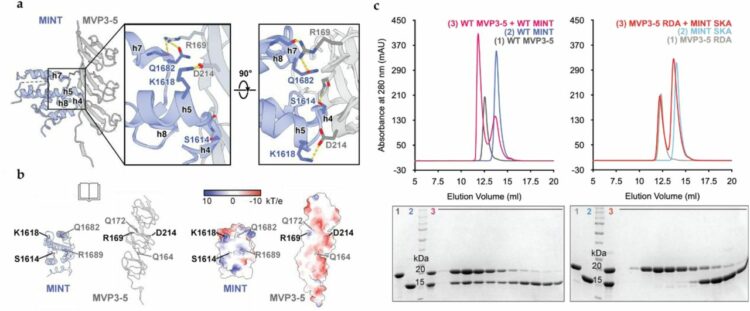
Lodwick, Jane E; Shen, Rong; Erramilli, Satchal; Xie, Yuan; Roganowicz, Karolina; Kossiakoff, Anthony A; Zhao, Minglei
Structural Insights into the Roles of PARP4 and NAD in the Human Vault Cage Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid38979142,
title = {Structural Insights into the Roles of PARP4 and NAD in the Human Vault Cage},
author = {Jane E Lodwick and Rong Shen and Satchal Erramilli and Yuan Xie and Karolina Roganowicz and Anthony A Kossiakoff and Minglei Zhao},
doi = {10.1101/2024.06.27.601040},
issn = {2692-8205},
year = {2024},
date = {2024-06-01},
urldate = {2024-06-01},
journal = {bioRxiv},
abstract = {Vault is a massive ribonucleoprotein complex found across Eukaryota. The major vault protein (MVP) oligomerizes into an ovular cage, which contains several minor vault components (MVCs) and is thought to transport transiently bound "cargo" molecules. Vertebrate vaults house a poly (ADP-ribose) polymerase (known as PARP4 in humans), which is the only MVC with known enzymatic activity. Despite being discovered decades ago, the molecular basis for PARP4's interaction with MVP remains unclear. In this study, we determined the structure of the human vault cage in complex with PARP4 and its enzymatic substrate NAD . The structures reveal atomic-level details of the protein-binding interface, as well as unexpected NAD -binding pockets within the interior of the vault cage. In addition, proteomics data show that human vaults purified from wild-type and PARP4-depleted cells interact with distinct subsets of proteins. Our results thereby support a model in which PARP4's specific incorporation into the vault cage helps to regulate vault's selection of cargo and its subcellular localization. Further, PARP4's proximity to MVP's NAD -binding sites could support its enzymatic function within the vault.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
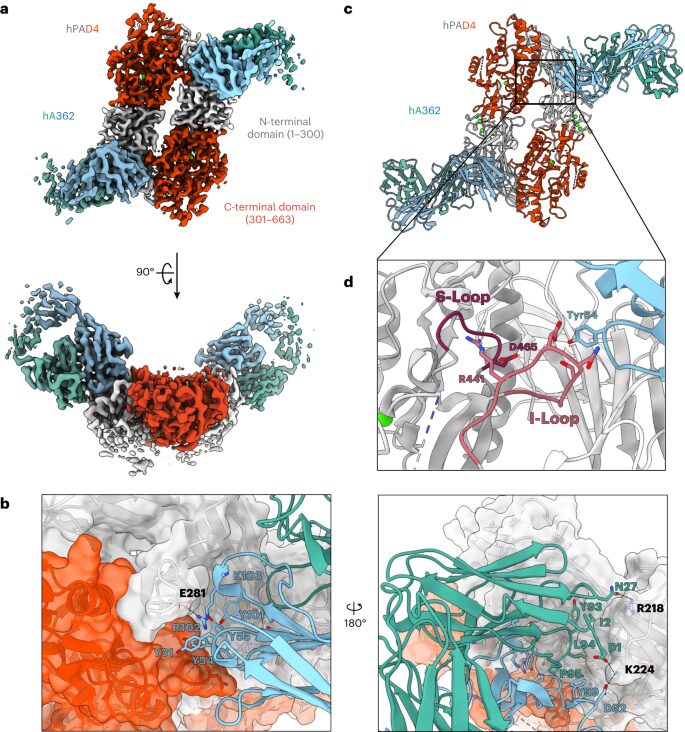
Zhou, Xin; Kong, Sophie; Maker, Allison; Remesh, Soumya G; Leung, Kevin K; Verba, Kliment A; Wells, James A
Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4 Journal Article
In: Nat Chem Biol, vol. 20, no. 6, pp. 742–750, 2024, ISSN: 1552-4469.
@article{pmid38308046,
title = {Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4},
author = {Xin Zhou and Sophie Kong and Allison Maker and Soumya G Remesh and Kevin K Leung and Kliment A Verba and James A Wells},
doi = {10.1038/s41589-023-01535-8},
issn = {1552-4469},
year = {2024},
date = {2024-06-01},
urldate = {2024-06-01},
journal = {Nat Chem Biol},
volume = {20},
number = {6},
pages = {742--750},
abstract = {Unlocking the potential of protein arginine deiminase 4 (PAD4) as a drug target for rheumatoid arthritis requires a deeper understanding of its regulation. In this study, we use unbiased antibody selections to identify functional antibodies capable of either activating or inhibiting PAD4 activity. Through cryogenic-electron microscopy, we characterized the structures of these antibodies in complex with PAD4 and revealed insights into their mechanisms of action. Rather than steric occlusion of the substrate-binding catalytic pocket, the antibodies modulate PAD4 activity through interactions with allosteric binding sites adjacent to the catalytic pocket. These binding events lead to either alteration of the active site conformation or the enzyme oligomeric state, resulting in modulation of PAD4 activity. Our study uses antibody engineering to reveal new mechanisms for enzyme regulation and highlights the potential of using PAD4 agonist and antagonist antibodies for studying PAD4-dependency in disease models and future therapeutic development.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
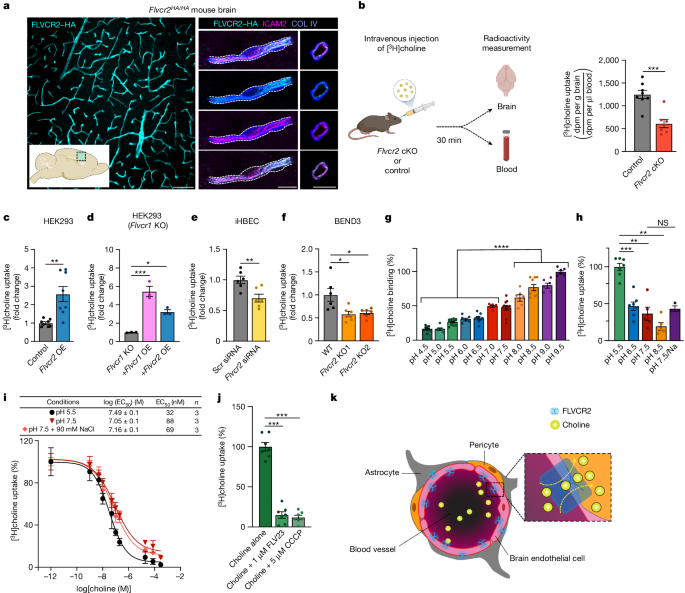
Cater, Rosemary J; Mukherjee, Dibyanti; Gil-Iturbe, Eva; Erramilli, Satchal K; Chen, Ting; Koo, Katie; Santander, Nicolás; Reckers, Andrew; Kloss, Brian; Gawda, Tomasz; Choy, Brendon C; Zhang, Zhening; Katewa, Aditya; Larpthaveesarp, Amara; Huang, Eric J; Mooney, Scott W J; Clarke, Oliver B; Yee, Sook Wah; Giacomini, Kathleen M; Kossiakoff, Anthony A; Quick, Matthias; Arnold, Thomas; Mancia, Filippo
Structural and molecular basis of choline uptake into the brain by FLVCR2 Journal Article
In: Nature, vol. 629, no. 8012, pp. 704–709, 2024, ISSN: 1476-4687.
@article{pmid38693257,
title = {Structural and molecular basis of choline uptake into the brain by FLVCR2},
author = {Rosemary J Cater and Dibyanti Mukherjee and Eva Gil-Iturbe and Satchal K Erramilli and Ting Chen and Katie Koo and Nicolás Santander and Andrew Reckers and Brian Kloss and Tomasz Gawda and Brendon C Choy and Zhening Zhang and Aditya Katewa and Amara Larpthaveesarp and Eric J Huang and Scott W J Mooney and Oliver B Clarke and Sook Wah Yee and Kathleen M Giacomini and Anthony A Kossiakoff and Matthias Quick and Thomas Arnold and Filippo Mancia},
doi = {10.1038/s41586-024-07326-y},
issn = {1476-4687},
year = {2024},
date = {2024-05-01},
urldate = {2024-05-01},
journal = {Nature},
volume = {629},
number = {8012},
pages = {704--709},
abstract = {Choline is an essential nutrient that the human body needs in vast quantities for cell membrane synthesis, epigenetic modification and neurotransmission. The brain has a particularly high demand for choline, but how it enters the brain remains unknown. The major facilitator superfamily transporter FLVCR1 (also known as MFSD7B or SLC49A1) was recently determined to be a choline transporter but is not highly expressed at the blood-brain barrier, whereas the related protein FLVCR2 (also known as MFSD7C or SLC49A2) is expressed in endothelial cells at the blood-brain barrier. Previous studies have shown that mutations in human Flvcr2 cause cerebral vascular abnormalities, hydrocephalus and embryonic lethality, but the physiological role of FLVCR2 is unknown. Here we demonstrate both in vivo and in vitro that FLVCR2 is a BBB choline transporter and is responsible for the majority of choline uptake into the brain. We also determine the structures of choline-bound FLVCR2 in both inward-facing and outward-facing states using cryo-electron microscopy. These results reveal how the brain obtains choline and provide molecular-level insights into how FLVCR2 binds choline in an aromatic cage and mediates its uptake. Our work could provide a novel framework for the targeted delivery of therapeutic agents into the brain.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Zhou, Jie; Le, Chau Q; Zhang, Yun; Wells, James A
A general approach for selection of epitope-directed binders to proteins Journal Article
In: Proc Natl Acad Sci U S A, vol. 121, no. 19, pp. e2317307121, 2024, ISSN: 1091-6490.
@article{pmid38683990,
title = {A general approach for selection of epitope-directed binders to proteins},
author = {Jie Zhou and Chau Q Le and Yun Zhang and James A Wells},
doi = {10.1073/pnas.2317307121},
issn = {1091-6490},
year = {2024},
date = {2024-05-01},
urldate = {2024-05-01},
journal = {Proc Natl Acad Sci U S A},
volume = {121},
number = {19},
pages = {e2317307121},
abstract = {Directing antibodies to a particular epitope among many possible on a target protein is a significant challenge. Here, we present a simple and general method for epitope-directed selection (EDS) using a differential phage selection strategy. This involves engineering the protein of interest (POI) with the epitope of interest (EOI) mutated using a systematic bioinformatics algorithm to guide the local design of an EOI decoy variant. Using several alternating rounds of negative selection with the EOI decoy variant followed by positive selection on the wild-type POI, we were able to identify highly specific and potent antibodies to five different EOI antigens that bind and functionally block known sites of proteolysis. Among these, we developed highly specific antibodies that target the proteolytic site on the CUB domain containing protein 1 (CDCP1) to prevent its proteolysis allowing us to study the cellular maturation of this event that triggers malignancy. We generated antibodies that recognize the junction between the pro- and catalytic domains for three different matrix metalloproteases (MMPs), MMP1, MMP3, and MMP9, that selectively block activation of each of these enzymes and impair cell migration. We targeted a proteolytic epitope on the cell surface receptor, EPH Receptor A2 (EphA2), that is known to transform it from a tumor suppressor to an oncoprotein. We believe that the EDS method greatly facilitates the generation of antibodies to specific EOIs on a wide range of proteins and enzymes for broad therapeutic and diagnostic applications.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Veggiani, Gianluca; Sidhu, Sachdev S
Generation and Selection of Synthetic Human Antibody Libraries via Phage Display Journal Article
In: Cold Spring Harb Protoc, vol. 2024, no. 4, pp. 108347, 2024, ISSN: 1559-6095.
@article{pmid37295821,
title = {Generation and Selection of Synthetic Human Antibody Libraries via Phage Display},
author = {Gianluca Veggiani and Sachdev S Sidhu},
doi = {10.1101/pdb.prot108347},
issn = {1559-6095},
year = {2024},
date = {2024-04-01},
urldate = {2024-04-01},
journal = {Cold Spring Harb Protoc},
volume = {2024},
number = {4},
pages = {108347},
abstract = {Synthetic antibody libraries enable the development of antibodies that can recognize virtually any antigen, with affinity and specificity profiles that are superior to those of natural antibodies. By using highly stable and optimized frameworks, synthetic antibody libraries can be rapidly generated by precisely designing synthetic DNA, allowing absolute control over the position and chemical diversity introduced while expanding the sequence space for antigen recognition. Here, we describe a detailed protocol for the generation of highly diverse synthetic antibody phage display libraries based on a single framework, with diversity genetically incorporated by using finely designed mutagenic oligonucleotides. This general method enables the facile construction of large antibody libraries with precisely tunable features, resulting in the rapid development of recombinant antibodies for virtually any antigen.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Xie, Xiao; Moon, Patrick J; Crossley, Steven W M; Bischoff, Amanda J; He, Dan; Li, Gen; Dao, Nam; Gonzalez-Valero, Angel; Reeves, Audrey G; McKenna, Jeffrey M; Elledge, Susanna K; Wells, James A; Toste, F Dean; Chang, Christopher J
Oxidative cyclization reagents reveal tryptophan cation-π interactions Journal Article
In: Nature, vol. 627, no. 8004, pp. 680–687, 2024, ISSN: 1476-4687.
@article{pmid38448587,
title = {Oxidative cyclization reagents reveal tryptophan cation-π interactions},
author = {Xiao Xie and Patrick J Moon and Steven W M Crossley and Amanda J Bischoff and Dan He and Gen Li and Nam Dao and Angel Gonzalez-Valero and Audrey G Reeves and Jeffrey M McKenna and Susanna K Elledge and James A Wells and F Dean Toste and Christopher J Chang},
doi = {10.1038/s41586-024-07140-6},
issn = {1476-4687},
year = {2024},
date = {2024-03-01},
urldate = {2024-03-01},
journal = {Nature},
volume = {627},
number = {8004},
pages = {680--687},
abstract = {Methods for selective covalent modification of amino acids on proteins can enable a diverse array of applications, spanning probes and modulators of protein function to proteomics. Owing to their high nucleophilicity, cysteine and lysine residues are the most common points of attachment for protein bioconjugation chemistry through acid-base reactivity. Here we report a redox-based strategy for bioconjugation of tryptophan, the rarest amino acid, using oxaziridine reagents that mimic oxidative cyclization reactions in indole-based alkaloid biosynthetic pathways to achieve highly efficient and specific tryptophan labelling. We establish the broad use of this method, termed tryptophan chemical ligation by cyclization (Trp-CLiC), for selectively appending payloads to tryptophan residues on peptides and proteins with reaction rates that rival traditional click reactions and enabling global profiling of hyper-reactive tryptophan sites across whole proteomes. Notably, these reagents reveal a systematic map of tryptophan residues that participate in cation-π interactions, including functional sites that can regulate protein-mediated phase-separation processes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Liu, Hongtao; Zakrzewicz, Dariusz; Nosol, Kamil; Irobalieva, Rossitza N; Mukherjee, Somnath; Bang-Sørensen, Rose; Goldmann, Nora; Kunz, Sebastian; Rossi, Lorenzo; Kossiakoff, Anthony A; Urban, Stephan; Glebe, Dieter; Geyer, Joachim; Locher, Kaspar P
Structure of antiviral drug bulevirtide bound to hepatitis B and D virus receptor protein NTCP Journal Article
In: Nat Commun, vol. 15, no. 1, pp. 2476, 2024, ISSN: 2041-1723.
@article{pmid38509088,
title = {Structure of antiviral drug bulevirtide bound to hepatitis B and D virus receptor protein NTCP},
author = {Hongtao Liu and Dariusz Zakrzewicz and Kamil Nosol and Rossitza N Irobalieva and Somnath Mukherjee and Rose Bang-Sørensen and Nora Goldmann and Sebastian Kunz and Lorenzo Rossi and Anthony A Kossiakoff and Stephan Urban and Dieter Glebe and Joachim Geyer and Kaspar P Locher},
doi = {10.1038/s41467-024-46706-w},
issn = {2041-1723},
year = {2024},
date = {2024-03-01},
urldate = {2024-03-01},
journal = {Nat Commun},
volume = {15},
number = {1},
pages = {2476},
abstract = {Cellular entry of the hepatitis B and D viruses (HBV/HDV) requires binding of the viral surface polypeptide preS1 to the hepatobiliary transporter Na-taurocholate co-transporting polypeptide (NTCP). This interaction can be blocked by bulevirtide (BLV, formerly Myrcludex B), a preS1 derivative and approved drug for treating HDV infection. Here, to elucidate the basis of this inhibitory function, we determined a cryo-EM structure of BLV-bound human NTCP. BLV forms two domains, a plug lodged in the bile salt transport tunnel of NTCP and a string that covers the receptor's extracellular surface. The N-terminally attached myristoyl group of BLV interacts with the lipid-exposed surface of NTCP. Our structure reveals how BLV inhibits bile salt transport, rationalizes NTCP mutations that decrease the risk of HBV/HDV infection, and provides a basis for understanding the host specificity of HBV/HDV. Our results provide opportunities for structure-guided development of inhibitors that target HBV/HDV docking to NTCP.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Nejo, Takahide; Wang, Lin; Leung, Kevin K; Wang, Albert; Lakshmanachetty, Senthilnath; Gallus, Marco; Kwok, Darwin W; Hong, Chibo; Chen, Lee H; Carrera, Diego A; Zhang, Michael Y; Stevers, Nicholas O; Maldonado, Gabriella C; Yamamichi, Akane; Watchmaker, Payal B; Naik, Akul; Shai, Anny; Phillips, Joanna J; Chang, Susan M; Wiita, Arun P; Wells, James A; Costello, Joseph F; Diaz, Aaron A; Okada, Hideho
Challenges in the discovery of tumor-specific alternative splicing-derived cell-surface antigens in glioma Journal Article
In: Sci Rep, vol. 14, no. 1, pp. 6362, 2024, ISSN: 2045-2322.
@article{pmid38493204,
title = {Challenges in the discovery of tumor-specific alternative splicing-derived cell-surface antigens in glioma},
author = {Takahide Nejo and Lin Wang and Kevin K Leung and Albert Wang and Senthilnath Lakshmanachetty and Marco Gallus and Darwin W Kwok and Chibo Hong and Lee H Chen and Diego A Carrera and Michael Y Zhang and Nicholas O Stevers and Gabriella C Maldonado and Akane Yamamichi and Payal B Watchmaker and Akul Naik and Anny Shai and Joanna J Phillips and Susan M Chang and Arun P Wiita and James A Wells and Joseph F Costello and Aaron A Diaz and Hideho Okada},
doi = {10.1038/s41598-024-56684-0},
issn = {2045-2322},
year = {2024},
date = {2024-03-01},
urldate = {2024-03-01},
journal = {Sci Rep},
volume = {14},
number = {1},
pages = {6362},
abstract = {Despite advancements in cancer immunotherapy, solid tumors remain formidable challenges. In glioma, profound inter- and intra-tumoral heterogeneity of antigen landscape hampers therapeutic development. Therefore, it is critical to consider alternative sources to expand the repertoire of targetable (neo-)antigens and improve therapeutic outcomes. Accumulating evidence suggests that tumor-specific alternative splicing (AS) could be an untapped reservoir of antigens. In this study, we investigated tumor-specific AS events in glioma, focusing on those predicted to generate major histocompatibility complex (MHC)-presentation-independent, cell-surface antigens that could be targeted by antibodies and chimeric antigen receptor-T cells. We systematically analyzed bulk RNA-sequencing datasets comparing 429 tumor samples (from The Cancer Genome Atlas) and 9166 normal tissue samples (from the Genotype-Tissue Expression project), and identified 13 AS events in 7 genes predicted to be expressed in more than 10% of the patients, including PTPRZ1 and BCAN, which were corroborated by an external RNA-sequencing dataset. Subsequently, we validated our predictions and elucidated the complexity of the isoforms using full-length transcript amplicon sequencing on patient-derived glioblastoma cells. However, analyses of the RNA-sequencing datasets of spatially mapped and longitudinally collected clinical tumor samples unveiled remarkable spatiotemporal heterogeneity of the candidate AS events. Furthermore, proteomics analysis did not reveal any peptide spectra matching the putative antigens. Our investigation illustrated the diverse characteristics of the tumor-specific AS events and the challenges of antigen exploration due to their notable spatiotemporal heterogeneity and elusive nature at the protein levels. Redirecting future efforts toward intracellular, MHC-presented antigens could offer a more viable avenue.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
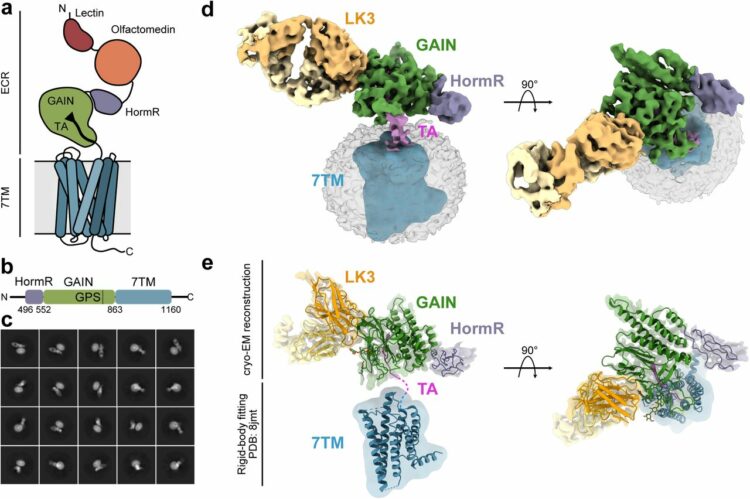
Kordon, Szymon P; Cechova, Kristina; Bandekar, Sumit J; Leon, Katherine; Dutka, Przemysław; Siffer, Gracie; Kossiakoff, Anthony A; Vafabakhsh, Reza; Araç, Demet
Structural analysis and conformational dynamics of a holo-adhesion GPCR reveal interplay between extracellular and transmembrane domains Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid38464178,
title = {Structural analysis and conformational dynamics of a holo-adhesion GPCR reveal interplay between extracellular and transmembrane domains},
author = {Szymon P Kordon and Kristina Cechova and Sumit J Bandekar and Katherine Leon and Przemysław Dutka and Gracie Siffer and Anthony A Kossiakoff and Reza Vafabakhsh and Demet Araç},
doi = {10.1101/2024.02.25.581807},
issn = {2692-8205},
year = {2024},
date = {2024-02-01},
urldate = {2024-02-01},
journal = {bioRxiv},
abstract = {Adhesion G Protein-Coupled Receptors (aGPCRs) are key cell-adhesion molecules involved in numerous physiological functions. aGPCRs have large multi-domain extracellular regions (ECR) containing a conserved GAIN domain that precedes their seven-pass transmembrane domain (7TM). Ligand binding and mechanical force applied on the ECR regulate receptor function. However, how the ECR communicates with the 7TM remains elusive, because the relative orientation and dynamics of the ECR and 7TM within a holoreceptor is unclear. Here, we describe the cryo-EM reconstruction of an aGPCR, Latrophilin3/ADGRL3, and reveal that the GAIN domain adopts a parallel orientation to the membrane and has constrained movement. Single-molecule FRET experiments unveil three slow-exchanging FRET states of the ECR relative to the 7TM within the holoreceptor. GAIN-targeted antibodies, and cancer-associated mutations at the GAIN-7TM interface, alter FRET states, cryo-EM conformations, and receptor signaling. Altogether, this data demonstrates conformational and functional coupling between the ECR and 7TM, suggesting an ECR-mediated mechanism of aGPCR activation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
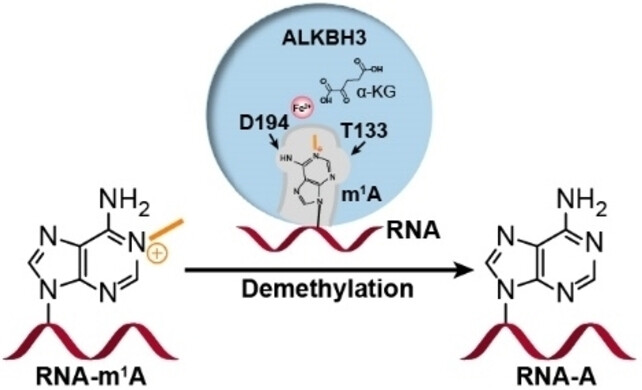
Zhang, Lin; Duan, Hong-Chao; Paduch, Marcin; Hu, Jingyan; Zhang, Chi; Mu, Yajuan; Lin, Houwen; He, Chuan; Kossiakoff, Anthony A; Jia, Guifang; Zhang, Liang
The Molecular Basis of Human ALKBH3 Mediated RNA N -methyladenosine (m A) Demethylation Journal Article
In: Angew Chem Int Ed Engl, vol. 63, no. 7, pp. e202313900, 2024, ISSN: 1521-3773.
@article{pmid38158383,
title = {The Molecular Basis of Human ALKBH3 Mediated RNA N -methyladenosine (m A) Demethylation},
author = {Lin Zhang and Hong-Chao Duan and Marcin Paduch and Jingyan Hu and Chi Zhang and Yajuan Mu and Houwen Lin and Chuan He and Anthony A Kossiakoff and Guifang Jia and Liang Zhang},
doi = {10.1002/anie.202313900},
issn = {1521-3773},
year = {2024},
date = {2024-02-01},
urldate = {2024-02-01},
journal = {Angew Chem Int Ed Engl},
volume = {63},
number = {7},
pages = {e202313900},
abstract = {N -methyladenosine (m A) is a prevalent post-transcriptional RNA modification, and the distribution and dynamics of the modification play key epitranscriptomic roles in cell development. At present, the human AlkB Fe(II)/α-ketoglutarate-dependent dioxygenase family member ALKBH3 is the only known mRNA m A demethylase, but its catalytic mechanism remains unclear. Here, we present the structures of ALKBH3-oligo crosslinked complexes obtained with the assistance of a synthetic antibody crystallization chaperone. Structural and biochemical results showed that ALKBH3 utilized two β-hairpins (β4-loop-β5 and β'-loop-β'') and the α2 helix to facilitate single-stranded substrate binding. Moreover, a bubble-like region around Asp194 and a key residue inside the active pocket (Thr133) enabled specific recognition and demethylation of m A- and 3-methylcytidine (m C)-modified substrates. Mutation of Thr133 to the corresponding residue in the AlkB Fe(II)/α-ketoglutarate-dependent dioxygenase family members FTO or ALKBH5 converted ALKBH3 substrate selectivity from m A to N -methyladenosine (m A), as did Asp194 deletion. Our findings provide a molecular basis for understanding the mechanisms of substrate recognition and m A demethylation by ALKBH3. This study is expected to aid structure-guided design of chemical probes for further functional studies and therapeutic applications.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
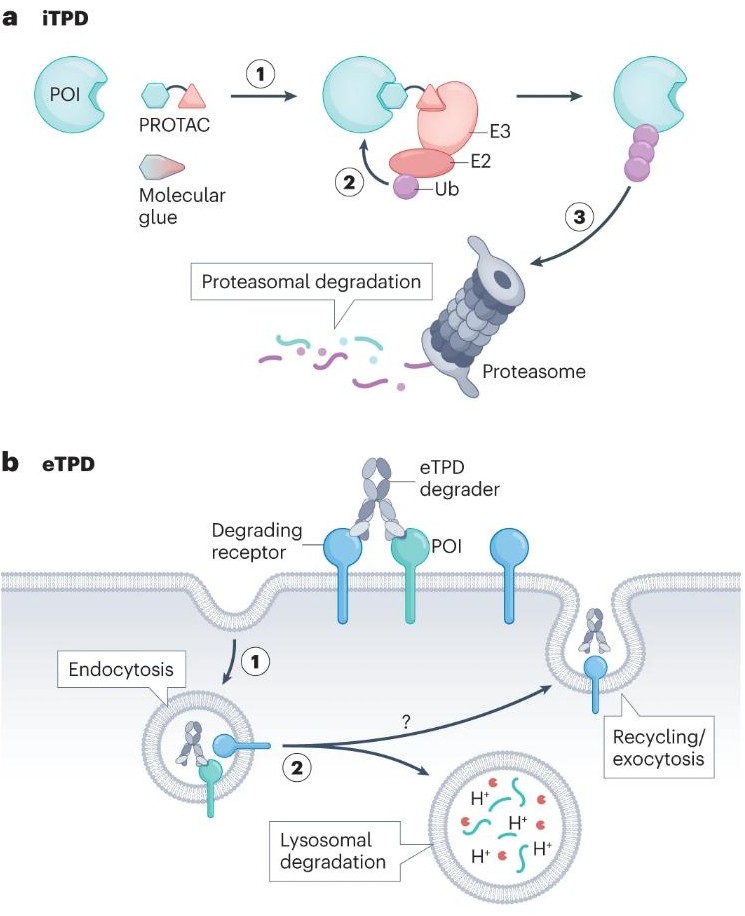
Wells, James A; Kumru, Kaan
Extracellular targeted protein degradation: an emerging modality for drug discovery Journal Article
In: Nat Rev Drug Discov, vol. 23, no. 2, pp. 126–140, 2024, ISSN: 1474-1784.
@article{pmid38062152,
title = {Extracellular targeted protein degradation: an emerging modality for drug discovery},
author = {James A Wells and Kaan Kumru},
doi = {10.1038/s41573-023-00833-z},
issn = {1474-1784},
year = {2024},
date = {2024-02-01},
urldate = {2024-02-01},
journal = {Nat Rev Drug Discov},
volume = {23},
number = {2},
pages = {126--140},
abstract = {Targeted protein degradation (TPD) has emerged in the past decade as a major new drug modality to remove intracellular proteins with bispecific small molecules that recruit the protein of interest (POI) to an E3 ligase for degradation in the proteasome. Unlike classic occupancy-based drugs, intracellular TPD (iTPD) eliminates the target and works catalytically, and so can be more effective and sustained, with lower dose requirements. Recently, this approach has been expanded to the extracellular proteome, including both secreted and membrane proteins. Extracellular targeted protein degradation (eTPD) uses bispecific antibodies, conjugates or small molecules to degrade extracellular POIs by trafficking them to the lysosome for degradation. Here, we focus on recent advances in eTPD, covering degrader systems, targets, molecular designs and parameters to advance them. Now almost any protein, intracellular or extracellular, is addressable in principle with TPD.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
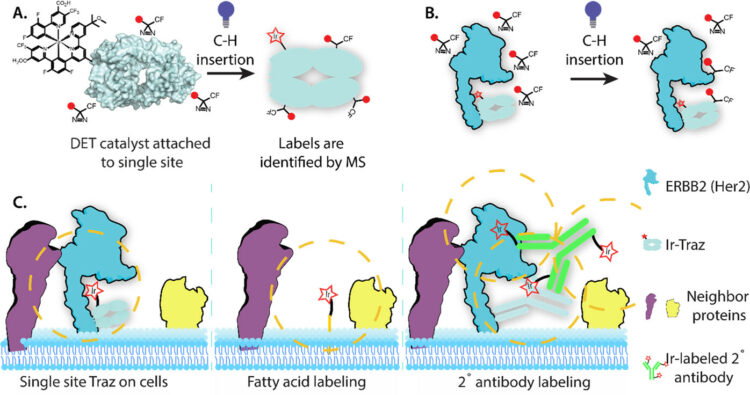
Bartholow, Thomas G; Burroughs, Paul W W; Elledge, Susanna K; Byrnes, James R; Kirkemo, Lisa L; Garda, Virginia; Leung, Kevin K; Wells, James A
In: ACS Cent Sci, vol. 10, no. 1, pp. 199–208, 2024, ISSN: 2374-7943.
@article{pmid38292613,
title = {Photoproximity Labeling from Single Catalyst Sites Allows Calibration and Increased Resolution for Carbene Labeling of Protein Partners In Vitro and on Cells},
author = {Thomas G Bartholow and Paul W W Burroughs and Susanna K Elledge and James R Byrnes and Lisa L Kirkemo and Virginia Garda and Kevin K Leung and James A Wells},
doi = {10.1021/acscentsci.3c01473},
issn = {2374-7943},
year = {2024},
date = {2024-01-01},
urldate = {2024-01-01},
journal = {ACS Cent Sci},
volume = {10},
number = {1},
pages = {199--208},
abstract = {The cell surface proteome (surfaceome) plays a pivotal role in virtually all extracellular biology, and yet we are only beginning to understand the protein complexes formed in this crowded environment. Recently, a high-resolution approach (μMap) was described that utilizes multiple iridium-photocatalysts attached to a secondary antibody, directed to a primary antibody of a protein of interest, to identify proximal neighbors by light-activated conversion of a biotin-diazirine to a highly reactive carbene followed by LC/MS (Geri, J. B.; Oakley, J. V.; Reyes-Robles, T.; Wang, T.; McCarver, S. J.; White, C. H.; Rodriguez-Rivera, F. P.; Parker, D. L.; Hett, E. C.; Fadeyi, O. O.; Oslund, R. C.; MacMillan, D. W. C. , , 1091-1097). Here we calibrated the spatial resolution for carbene labeling using site-specific conjugation of a single photocatalyst to a primary antibody drug, trastuzumab (Traz), in complex with its structurally well-characterized oncogene target, HER2. We observed relatively uniform carbene labeling across all amino acids, and a maximum distance of ∼110 Å from the fixed photocatalyst. When targeting HER2 overexpression cells, we identified 20 highly enriched HER2 neighbors, compared to a nonspecific membrane tethered catalyst. These studies identify new HER2 interactors and calibrate the radius of carbene photoprobe labeling for the surfaceome.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
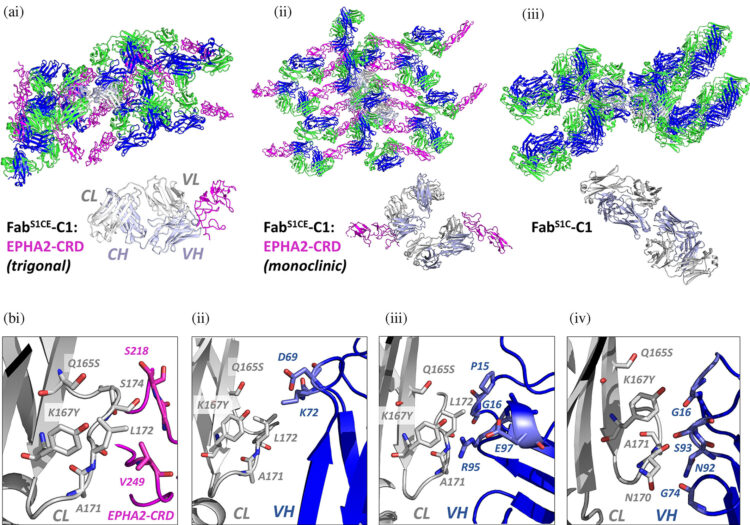
Bruce, Heather A; Singer, Alexander U; Filippova, Ekaterina V; Blazer, Levi L; Adams, Jarrett J; Enderle, Leonie; Ben-David, Moshe; Radley, Elizabeth H; Mao, Daniel Y L; Pau, Victor; Orlicky, Stephen; Sicheri, Frank; Kurinov, Igor; Atwell, Shane; Kossiakoff, Anthony A; Sidhu, Sachdev S
Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes Journal Article
In: Protein Sci, vol. 33, no. 1, pp. e4824, 2024, ISSN: 1469-896X.
@article{pmid37945533,
title = {Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes},
author = {Heather A Bruce and Alexander U Singer and Ekaterina V Filippova and Levi L Blazer and Jarrett J Adams and Leonie Enderle and Moshe Ben-David and Elizabeth H Radley and Daniel Y L Mao and Victor Pau and Stephen Orlicky and Frank Sicheri and Igor Kurinov and Shane Atwell and Anthony A Kossiakoff and Sachdev S Sidhu},
doi = {10.1002/pro.4824},
issn = {1469-896X},
year = {2024},
date = {2024-01-01},
urldate = {2024-01-01},
journal = {Protein Sci},
volume = {33},
number = {1},
pages = {e4824},
abstract = {The atomic-resolution structural information that X-ray crystallography can provide on the binding interface between a Fab and its cognate antigen is highly valuable for understanding the mechanism of interaction. However, many Fab:antigen complexes are recalcitrant to crystallization, making the endeavor a considerable effort with no guarantee of success. Consequently, there have been significant steps taken to increase the likelihood of Fab:antigen complex crystallization by altering the Fab framework. In this investigation, we applied the surface entropy reduction strategy coupled with phage-display technology to identify a set of surface substitutions that improve the propensity of a human Fab framework to crystallize. In addition, we showed that combining these surface substitutions with previously reported Crystal Kappa and elbow substitutions results in an extraordinary improvement in Fab and Fab:antigen complex crystallizability, revealing a strong synergistic relationship between these sets of substitutions. Through comprehensive Fab and Fab:antigen complex crystallization screenings followed by structure determination and analysis, we defined the roles that each of these substitutions play in facilitating crystallization and how they complement each other in the process.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
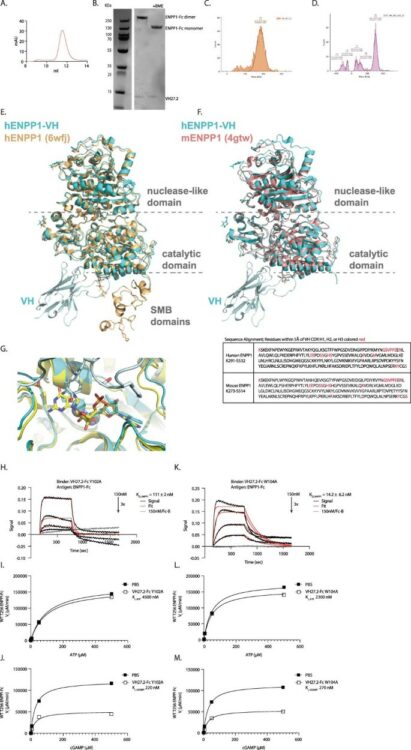
Solomon, Paige E; Bracken, Colton J; Carozza, Jacqueline A; Wang, Haoqing; Young, Elizabeth P; Wellner, Alon; Liu, Chang C; Sweet-Cordero, E Alejandro; Li, Lingyin; Wells, James A
Discovery of VH domains that allosterically inhibit ENPP1 Journal Article
In: Nat Chem Biol, vol. 20, no. 1, pp. 30–41, 2024, ISSN: 1552-4469.
@article{pmid37400538,
title = {Discovery of VH domains that allosterically inhibit ENPP1},
author = {Paige E Solomon and Colton J Bracken and Jacqueline A Carozza and Haoqing Wang and Elizabeth P Young and Alon Wellner and Chang C Liu and E Alejandro Sweet-Cordero and Lingyin Li and James A Wells},
doi = {10.1038/s41589-023-01368-5},
issn = {1552-4469},
year = {2024},
date = {2024-01-01},
urldate = {2024-01-01},
journal = {Nat Chem Biol},
volume = {20},
number = {1},
pages = {30--41},
abstract = {Ectodomain phosphatase/phosphodiesterase-1 (ENPP1) is overexpressed on cancer cells and functions as an innate immune checkpoint by hydrolyzing extracellular cyclic guanosine monophosphate adenosine monophosphate (cGAMP). Biologic inhibitors have not yet been reported and could have substantial therapeutic advantages over current small molecules because they can be recombinantly engineered into multifunctional formats and immunotherapies. Here we used phage and yeast display coupled with in cellulo evolution to generate variable heavy (VH) single-domain antibodies against ENPP1 and discovered a VH domain that allosterically inhibited the hydrolysis of cGAMP and adenosine triphosphate (ATP). We solved a 3.2 Å-resolution cryo-electron microscopy structure for the VH inhibitor complexed with ENPP1 that confirmed its new allosteric binding pose. Finally, we engineered the VH domain into multispecific formats and immunotherapies, including a bispecific fusion with an anti-PD-L1 checkpoint inhibitor that showed potent cellular activity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}