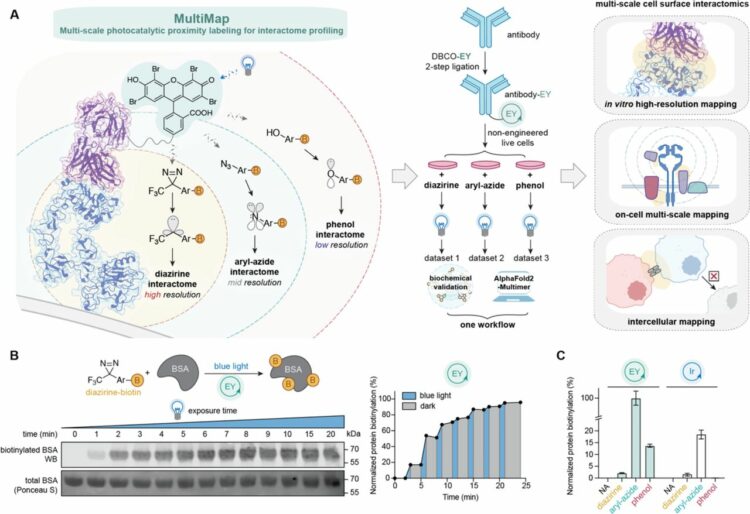
Publications: 2023
2023
Lin, Zhi; Schaefer, Kaitlin; Lui, Irene; Yao, Zi; Fossati, Andrea; Swaney, Danielle L; Palar, Ajikarunia; Sali, Andrej; Wells, James A
Multi-scale photocatalytic proximity labeling reveals cell surface neighbors on and between cells Journal Article
In: bioRxiv, 2023, ISSN: 2692-8205.
@article{pmid37961561,
title = {Multi-scale photocatalytic proximity labeling reveals cell surface neighbors on and between cells},
author = {Zhi Lin and Kaitlin Schaefer and Irene Lui and Zi Yao and Andrea Fossati and Danielle L Swaney and Ajikarunia Palar and Andrej Sali and James A Wells},
doi = {10.1101/2023.10.28.564055},
issn = {2692-8205},
year = {2023},
date = {2023-10-01},
urldate = {2023-10-01},
journal = {bioRxiv},
abstract = {The cell membrane proteome is the primary biohub for cell communication, yet we are only beginning to understand the dynamic protein neighborhoods that form on the cell surface and between cells. Proximity labeling proteomics (PLP) strategies using chemically reactive probes are powerful approaches to yield snapshots of protein neighborhoods but are currently limited to one single resolution based on the probe labeling radius. Here, we describe a multi-scale PLP method with tunable resolution using a commercially available histological dye, Eosin Y, which upon visible light illumination, activates three different photo-probes with labeling radii ranging from ∼100 to 3000 Å. We applied this platform to profile neighborhoods of the oncogenic epidermal growth factor receptor (EGFR) and orthogonally validated >20 neighbors using immuno-assays and AlphaFold-Multimer prediction that generated plausible binary interaction models. We further profiled the protein neighborhoods of cell-cell synapses induced by bi-specific T-cell engagers (BiTEs) and chimeric antigen receptor (CAR)T cells at longer length scales. This integrated multi-scale PLP platform maps local and distal protein networks on cell surfaces and between cells. We believe this information will aid in the systematic construction of the cell surface interactome and reveal new opportunities for immunotherapeutics.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
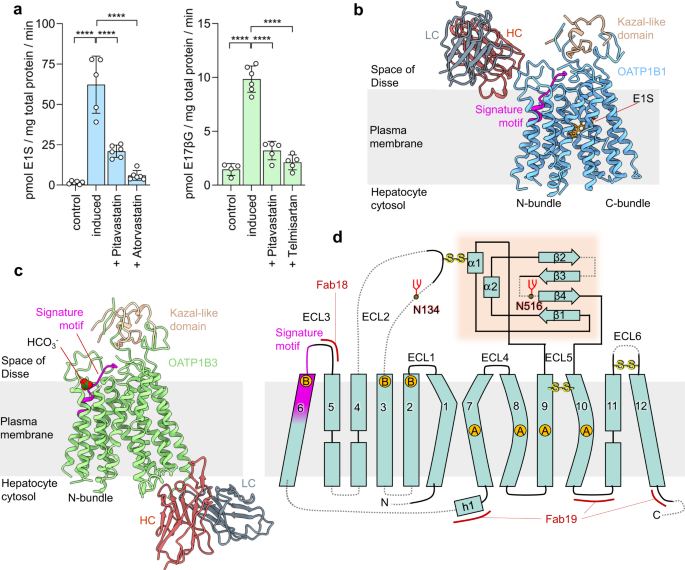
Ciută, Anca-Denise; Nosol, Kamil; Kowal, Julia; Mukherjee, Somnath; Ramírez, Ana S; Stieger, Bruno; Kossiakoff, Anthony A; Locher, Kaspar P
Structure of human drug transporters OATP1B1 and OATP1B3 Journal Article
In: Nat Commun, vol. 14, no. 1, pp. 5774, 2023, ISSN: 2041-1723.
@article{pmid37723174,
title = {Structure of human drug transporters OATP1B1 and OATP1B3},
author = {Anca-Denise Ciută and Kamil Nosol and Julia Kowal and Somnath Mukherjee and Ana S Ramírez and Bruno Stieger and Anthony A Kossiakoff and Kaspar P Locher},
doi = {10.1038/s41467-023-41552-8},
issn = {2041-1723},
year = {2023},
date = {2023-09-01},
urldate = {2023-09-01},
journal = {Nat Commun},
volume = {14},
number = {1},
pages = {5774},
abstract = {The organic anion transporting polypeptides OATP1B1 and OATP1B3 are membrane proteins that mediate uptake of drugs into the liver for subsequent conjugation and biliary excretion, a key step in drug elimination from the human body. Polymorphic variants of these transporters can cause reduced drug clearance and adverse drug effects such as statin-induced rhabdomyolysis, and co-administration of OATP substrates can lead to damaging drug-drug interaction. Despite their clinical relevance in drug disposition and pharmacokinetics, the structure and mechanism of OATPs are unknown. Here we present cryo-EM structures of human OATP1B1 and OATP1B3 bound to synthetic Fab fragments and in functionally distinct states. A single estrone-3-sulfate molecule is bound in a pocket located in the C-terminal half of OATP1B1. The shape and chemical nature of the pocket rationalize the preference for diverse organic anions and allow in silico docking of statins. The structure of OATP1B3 is determined in a drug-free state but reveals a bicarbonate molecule bound to the conserved signature motif and a histidine residue that is prevalent in OATPs exhibiting pH-dependent activity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
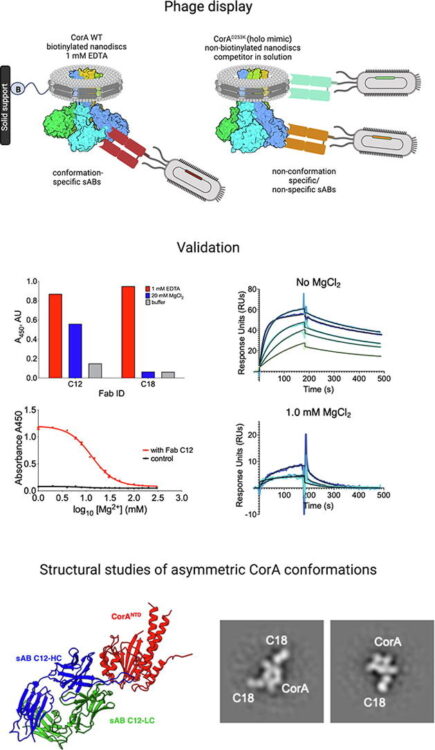
Erramilli, Satchal K; Dominik, Pawel K; Deneka, Dawid; Tokarz, Piotr; Kim, Sangwoo S; Reddy, Bharat G; Skrobek, Blazej M; Dalmas, Olivier; Perozo, Eduardo; Kossiakoff, Anthony A
Conformation-specific Synthetic Antibodies Discriminate Multiple Functional States of the Ion Channel CorA Journal Article
In: J Mol Biol, vol. 435, no. 17, pp. 168192, 2023, ISSN: 1089-8638.
@article{pmid37394032,
title = {Conformation-specific Synthetic Antibodies Discriminate Multiple Functional States of the Ion Channel CorA},
author = {Satchal K Erramilli and Pawel K Dominik and Dawid Deneka and Piotr Tokarz and Sangwoo S Kim and Bharat G Reddy and Blazej M Skrobek and Olivier Dalmas and Eduardo Perozo and Anthony A Kossiakoff},
doi = {10.1016/j.jmb.2023.168192},
issn = {1089-8638},
year = {2023},
date = {2023-09-01},
urldate = {2023-09-01},
journal = {J Mol Biol},
volume = {435},
number = {17},
pages = {168192},
abstract = {CorA, the primary magnesium ion channel in prokaryotes and archaea, is a prototypical homopentameric ion channel that undergoes ion-dependent conformational transitions. CorA adopts five-fold symmetric non-conductive states in the presence of high concentrations of Mg, and highly asymmetric flexible states in its complete absence. However, the latter were of insufficient resolution to be thoroughly characterized. In order to gain additional insights into the relationship between asymmetry and channel activation, we exploited phage display selection strategies to generate conformation-specific synthetic antibodies (sABs) against CorA in the absence of Mg. Two sABs from these selections, C12 and C18, showed different degrees of Mg-sensitivity. Through structural, biochemical, and biophysical characterization, we found the sABs are both conformation-specific but probe different features of the channel under open-like conditions. C18 is highly specific to the Mg-depleted state of CorA and through negative-stain electron microscopy (ns-EM), we show sAB binding reflects the asymmetric arrangement of CorA protomers in Mg-depleted conditions. We used X-ray crystallography to determine a structure at 2.0 Å resolution of sAB C12 bound to the soluble N-terminal regulatory domain of CorA. The structure shows C12 is a competitive inhibitor of regulatory magnesium binding through its interaction with the divalent cation sensing site. We subsequently exploited this relationship to capture and visualize asymmetric CorA states in different [Mg] using ns-EM. We additionally utilized these sABs to provide insights into the energy landscape that governs the ion-dependent conformational transitions of CorA.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
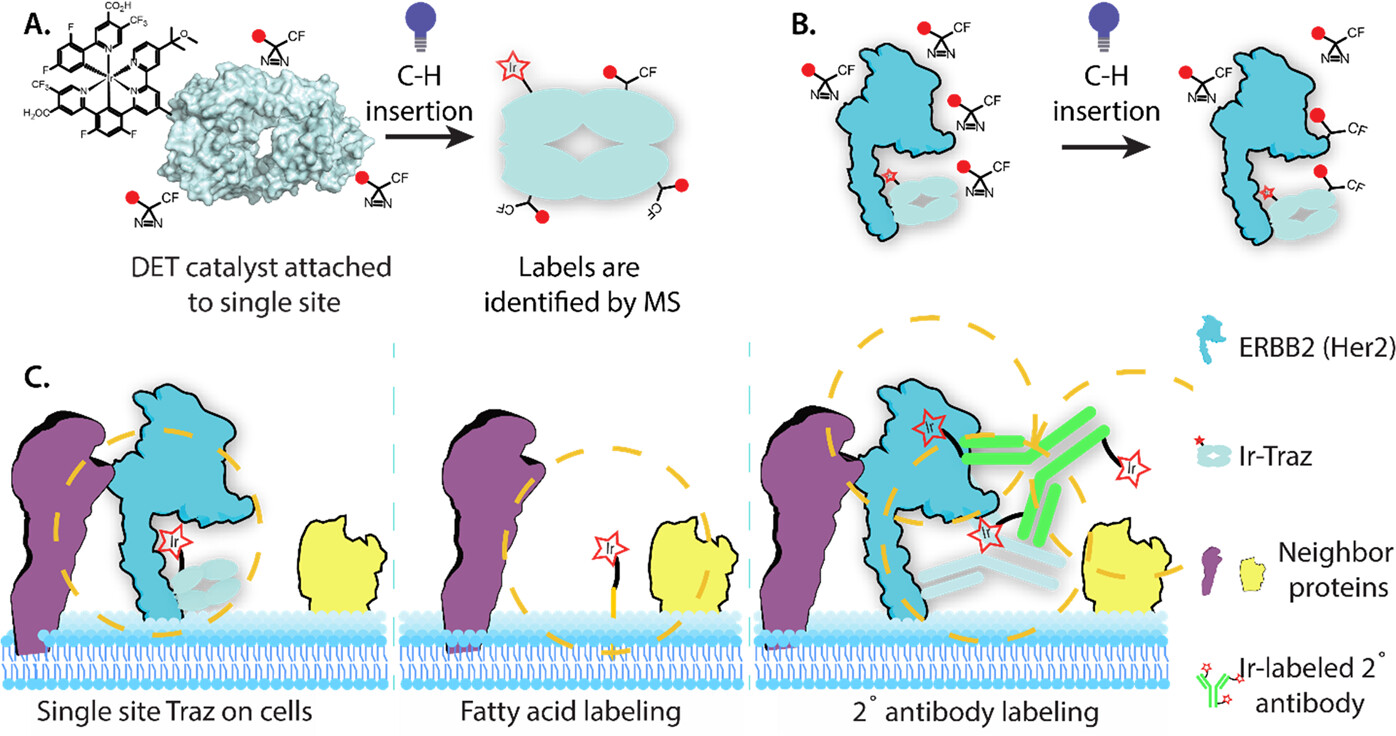
Bartholow, Thomas G; Burroughs, Paul; Elledge, Susanna K; Byrnes, James R; Kirkemo, Lisa L; Garda, Virginia; Leung, Kevin K; Wells, James A
Site-specific proximity labeling at single residue resolution for identification of protein partners and on cells Journal Article
In: bioRxiv, 2023, ISSN: 2692-8205.
@article{pmid37546992,
title = {Site-specific proximity labeling at single residue resolution for identification of protein partners and on cells},
author = {Thomas G Bartholow and Paul Burroughs and Susanna K Elledge and James R Byrnes and Lisa L Kirkemo and Virginia Garda and Kevin K Leung and James A Wells},
doi = {10.1101/2023.07.27.550738},
issn = {2692-8205},
year = {2023},
date = {2023-07-01},
urldate = {2023-07-01},
journal = {bioRxiv},
abstract = {The cell surface proteome, or surfaceome, is encoded by more than 4000 genes, but we are only beginning to understand the complexes they form. Rapid proximity labeling around specific membrane targets allows for capturing weak and transient interactions expected in the crowded and dynamic environment of the surfaceome. Recently, a high-resolution approach called μMap has been described (Geri, J. B., Oakley, J. V., Reyes-Robles, T., Wang, T., McCarver, S. J., White, C. H., Rodriguez-Rivera, F. P., Parker, D. L., Hett, E. C., Fadeyi, O. O., Oslund, R. C., and MacMillan, D. W. C. (2020) , 1091-1097) in which an iridium (Ir)-based photocatalyst is attached to a specific antibody to target labeling of neighbors utilizing light-activated generation of carbenes from diazirine compounds via Dexter Energy Transfer (DET). Here we studied and optimized the spatial resolution for the method using an oncoprotein complex between the antibody drug, trastuzumab (Traz), and its target HER2. A set of eight single site-specific Ir-catalytic centers were engineered into Traz to study intra- and inter-molecular labeling and on cells by mass spectrometry. From this structurally well-characterized complex we observed a maximum distance of ∼110 Å for labeling. Labeling occurred almost uniformly over the full range of amino acids, unlike the residue specific labeling of other techniques. To examine on cell labeling that is specific to HER2 as opposed to simply being on the membrane, we compared the labeling patterns for the eight Traz-catalyst species to random labeling of membrane proteins using a metabolically integrated fatty acid catalyst. Our results identified 20 high confidence HER2 neighbors, many novel, that were more than 6-fold enriched compared to the non-specific membrane tethered catalyst. These studies define distance labeling parameters from single-site catalysts placed directly on the membrane target of interest, and more accurately compare to non-specific labeling to identify membrane complexes with higher confidence.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
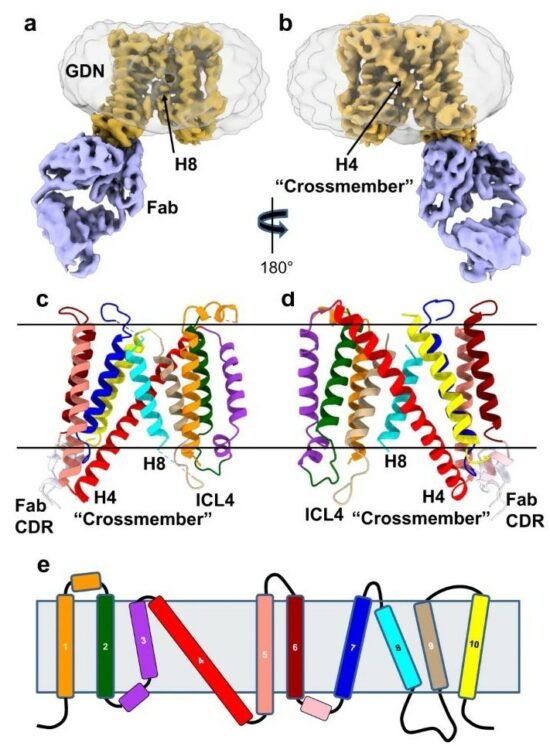
Leonhardt, Susan A; Purdy, Michael D; Grover, Jonathan R; Yang, Ziwei; Poulos, Sandra; McIntire, William E; Tatham, Elizabeth A; Erramilli, Satchal K; Nosol, Kamil; Lai, Kin Kui; Ding, Shilei; Lu, Maolin; Uchil, Pradeep D; Finzi, Andrés; Rein, Alan; Kossiakoff, Anthony A; Mothes, Walther; Yeager, Mark
Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry Journal Article
In: Nat Commun, vol. 14, no. 1, pp. 4368, 2023, ISSN: 2041-1723.
@article{pmid37474505,
title = {Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry},
author = {Susan A Leonhardt and Michael D Purdy and Jonathan R Grover and Ziwei Yang and Sandra Poulos and William E McIntire and Elizabeth A Tatham and Satchal K Erramilli and Kamil Nosol and Kin Kui Lai and Shilei Ding and Maolin Lu and Pradeep D Uchil and Andrés Finzi and Alan Rein and Anthony A Kossiakoff and Walther Mothes and Mark Yeager},
doi = {10.1038/s41467-023-39262-2},
issn = {2041-1723},
year = {2023},
date = {2023-07-01},
urldate = {2023-07-01},
journal = {Nat Commun},
volume = {14},
number = {1},
pages = {4368},
abstract = {The host proteins SERINC3 and SERINC5 are HIV-1 restriction factors that reduce infectivity when incorporated into the viral envelope. The HIV-1 accessory protein Nef abrogates incorporation of SERINCs via binding to intracellular loop 4 (ICL4). Here, we determine cryoEM maps of full-length human SERINC3 and an ICL4 deletion construct, which reveal that hSERINC3 is comprised of two α-helical bundles connected by a ~ 40-residue, highly tilted, "crossmember" helix. The design resembles non-ATP-dependent lipid transporters. Consistently, purified hSERINCs reconstituted into proteoliposomes induce flipping of phosphatidylserine (PS), phosphatidylethanolamine and phosphatidylcholine. Furthermore, SERINC3, SERINC5 and the scramblase TMEM16F expose PS on the surface of HIV-1 and reduce infectivity, with similar results in MLV. SERINC effects in HIV-1 and MLV are counteracted by Nef and GlycoGag, respectively. Our results demonstrate that SERINCs are membrane transporters that flip lipids, resulting in a loss of membrane asymmetry that is strongly correlated with changes in Env conformation and loss of infectivity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
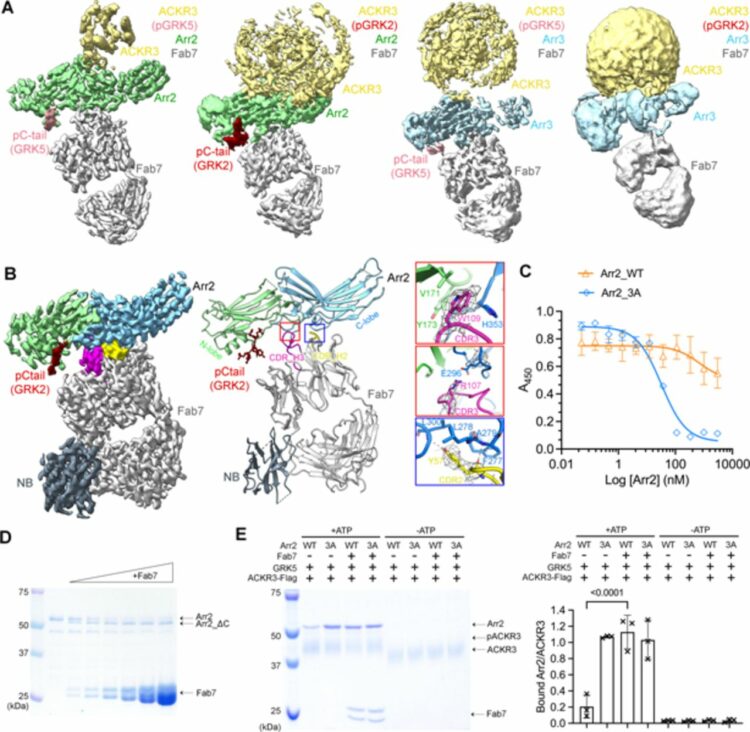
Chen, Qiuyan; Schafer, Christopher T; Mukherjee, Somnath; Gustavsson, Martin; Agrawal, Parth; Yao, Xin-Qiu; Kossiakoff, Anthony A; Handel, Tracy M; Tesmer, John J G
ACKR3-arrestin2/3 complexes reveal molecular consequences of GRK-dependent barcoding Journal Article
In: bioRxiv, 2023, ISSN: 2692-8205.
@article{pmid37502840,
title = {ACKR3-arrestin2/3 complexes reveal molecular consequences of GRK-dependent barcoding},
author = {Qiuyan Chen and Christopher T Schafer and Somnath Mukherjee and Martin Gustavsson and Parth Agrawal and Xin-Qiu Yao and Anthony A Kossiakoff and Tracy M Handel and John J G Tesmer},
doi = {10.1101/2023.07.18.549504},
issn = {2692-8205},
year = {2023},
date = {2023-07-01},
urldate = {2023-07-01},
journal = {bioRxiv},
abstract = {Atypical chemokine receptor 3 (ACKR3, also known as CXCR7) is a scavenger receptor that regulates extracellular levels of the chemokine CXCL12 to maintain responsiveness of its partner, the G protein-coupled receptor (GPCR), CXCR4. ACKR3 is notable because it does not couple to G proteins and instead is completely biased towards arrestins. Our previous studies revealed that GRK2 and GRK5 install distinct distributions of phosphates (or "barcodes") on the ACKR3 carboxy terminal tail, but how these unique barcodes drive different cellular outcomes is not understood. It is also not known if arrestin2 (Arr2) and 3 (Arr3) bind to these barcodes in distinct ways. Here we report cryo-electron microscopy structures of Arr2 and Arr3 in complex with ACKR3 phosphorylated by either GRK2 or GRK5. Unexpectedly, the finger loops of Arr2 and 3 directly insert into the detergent/membrane instead of the transmembrane core of ACKR3, in contrast to previously reported "core" GPCR-arrestin complexes. The distance between the phosphorylation barcode and the receptor transmembrane core regulates the interaction mode of arrestin, alternating between a tighter complex for GRK5 sites and heterogenous primarily "tail only" complexes for GRK2 sites. Arr2 and 3 bind at different angles relative to the core of ACKR3, likely due to differences in membrane/micelle anchoring at their C-edge loops. Our structural investigations were facilitated by Fab7, a novel Fab that binds both Arr2 and 3 in their activated states irrespective of receptor or phosphorylation status, rendering it a potentially useful tool to aid structure determination of any native GPCR-arrestin complex. The structures provide unprecedented insight into how different phosphorylation barcodes and arrestin isoforms can globally affect the configuration of receptor-arrestin complexes. These differences may promote unique downstream intracellular interactions and cellular responses. Our structures also suggest that the 100% bias of ACKR3 for arrestins is driven by the ability of arrestins, but not G proteins, to bind GRK-phosphorylated ACKR3 even when excluded from the receptor cytoplasmic binding pocket.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
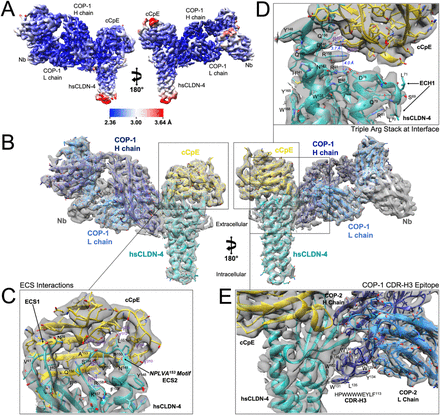
Erramilli, Satchal K; Dominik, Pawel K; Ogbu, Chinemerem P; Kossiakoff, Anthony A; Vecchio, Alex J
Cryo-EM structures of a synthetic antibody against 22 kDa claudin-4 reveal its complex with enterotoxin Journal Article
In: bioRxiv, 2023, ISSN: 2692-8205.
@article{pmid37398044,
title = {Cryo-EM structures of a synthetic antibody against 22 kDa claudin-4 reveal its complex with enterotoxin},
author = {Satchal K Erramilli and Pawel K Dominik and Chinemerem P Ogbu and Anthony A Kossiakoff and Alex J Vecchio},
doi = {10.1101/2023.06.12.544689},
issn = {2692-8205},
year = {2023},
date = {2023-06-01},
urldate = {2023-06-01},
journal = {bioRxiv},
abstract = {Claudins are a family of ∼25 kDa membrane proteins that integrate into tight junctions to form molecular barriers at the paracellular spaces between endothelial and epithelial cells. Humans have 27 subtypes, which homo- and hetero-oligomerize to impart distinct properties and physiological functions to tissues and organs. As the structural and functional backbone of tight junctions, claudins are attractive targets for therapeutics capable of modulating tissue permeability to deliver drugs or treat disease. However, structures of claudins are limited due to their small sizes and physicochemical properties-these traits also make therapy development a challenge. We have developed a synthetic antibody fragment (sFab) that binds human claudin-4 and used it to resolve structures of its complex with enterotoxin (CpE) using cryogenic electron microscopy (cryo-EM). The resolution of the structures reveals the architectures of 22 kDa claudin-4, the 14 kDa C-terminal domain of CpE, and the mechanism by which this sFab binds claudins. Further, we elucidate the biochemical and biophysical bases of sFab binding and demonstrate that this molecule exhibits subtype-selectivity by assaying homologous claudins. Our results provide a framework for developing sFabs against hard-to-target claudins and establishes the utility of sFabs as fiducial markers for determining cryo-EM structures of this small membrane protein family at resolutions that surpass X-ray crystallography. Taken together, this work highlights the ability of sFabs to elucidate claudin structure and function and posits their potential as therapeutics for modulating tight junctions by targeting specific claudin subtypes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
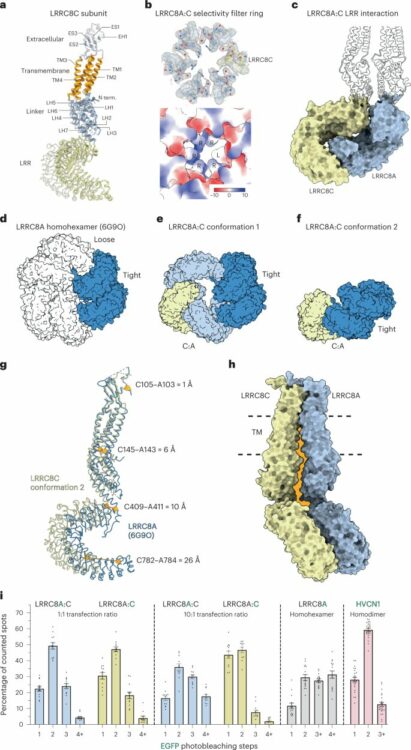
Kern, David M; Bleier, Julia; Mukherjee, Somnath; Hill, Jennifer M; Kossiakoff, Anthony A; Isacoff, Ehud Y; Brohawn, Stephen G
Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels Journal Article
In: Nat Struct Mol Biol, vol. 30, no. 6, pp. 841–852, 2023, ISSN: 1545-9985.
@article{pmid36928458,
title = {Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels},
author = {David M Kern and Julia Bleier and Somnath Mukherjee and Jennifer M Hill and Anthony A Kossiakoff and Ehud Y Isacoff and Stephen G Brohawn},
doi = {10.1038/s41594-023-00944-6},
issn = {1545-9985},
year = {2023},
date = {2023-06-01},
urldate = {2023-06-01},
journal = {Nat Struct Mol Biol},
volume = {30},
number = {6},
pages = {841--852},
abstract = {Leucine-rich repeat-containing protein 8 (LRRC8) family members form volume-regulated anion channels activated by hypoosmotic cell swelling. LRRC8 channels are ubiquitously expressed in vertebrate cells as heteromeric assemblies of LRRC8A (SWELL1) and LRRC8B-E subunits. Channels of different subunit composition have distinct properties that explain the functional diversity of LRRC8 currents across cell types. However, the basis for heteromeric LRRC8 channel assembly and function is unknown. Here we leverage a fiducial-tagging strategy to determine single-particle cryo-EM structures of heterohexameric LRRC8A:C channels in multiple conformations. Compared to homomers, LRRC8A:C channels show pronounced differences in architecture due to heterotypic LRR interactions that displace subunits away from the conduction axis and poise the channel for activation. Structures and functional studies further reveal that lipids embedded in the channel pore block ion conduction in the closed state. These results provide insight into determinants for heteromeric LRRC8 channel assembly, activity and gating by lipids.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
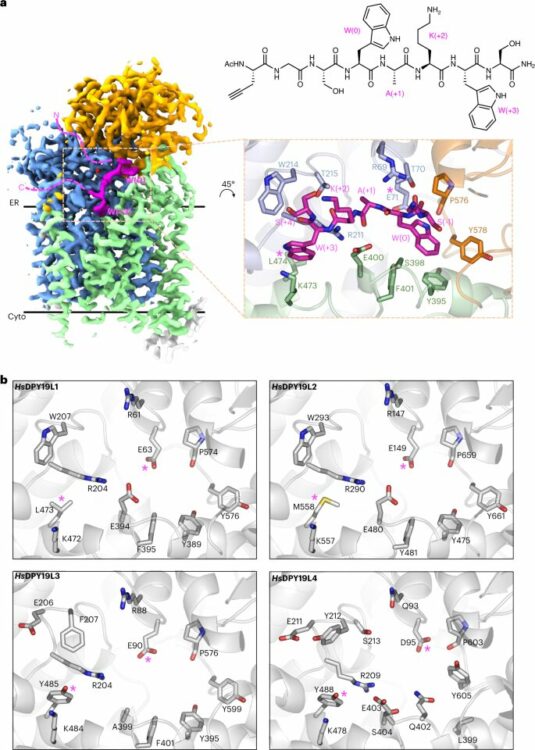
Bloch, Joël S; John, Alan; Mao, Runyu; Mukherjee, Somnath; Boilevin, Jérémy; Irobalieva, Rossitza N; Darbre, Tamis; Scott, Nichollas E; Reymond, Jean-Louis; Kossiakoff, Anthony A; Goddard-Borger, Ethan D; Locher, Kaspar P
Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase Journal Article
In: Nat Chem Biol, vol. 19, no. 5, pp. 575–584, 2023, ISSN: 1552-4469.
@article{pmid36604564,
title = {Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase},
author = {Joël S Bloch and Alan John and Runyu Mao and Somnath Mukherjee and Jérémy Boilevin and Rossitza N Irobalieva and Tamis Darbre and Nichollas E Scott and Jean-Louis Reymond and Anthony A Kossiakoff and Ethan D Goddard-Borger and Kaspar P Locher},
doi = {10.1038/s41589-022-01219-9},
issn = {1552-4469},
year = {2023},
date = {2023-05-01},
urldate = {2023-05-01},
journal = {Nat Chem Biol},
volume = {19},
number = {5},
pages = {575--584},
abstract = {C-linked glycosylation is essential for the trafficking, folding and function of secretory and transmembrane proteins involved in cellular communication processes. The tryptophan C-mannosyltransferase (CMT) enzymes that install the modification attach a mannose to the first tryptophan of WxxW/C sequons in nascent polypeptide chains by an unknown mechanism. Here, we report cryogenic-electron microscopy structures of Caenorhabditis elegans CMT in four key states: apo, acceptor peptide-bound, donor-substrate analog-bound and as a trapped ternary complex with both peptide and a donor-substrate mimic bound. The structures indicate how the C-mannosylation sequon is recognized by this CMT and its paralogs, and how sequon binding triggers conformational activation of the donor substrate: a process relevant to all glycosyltransferase C superfamily enzymes. Our structural data further indicate that the CMTs adopt an unprecedented electrophilic aromatic substitution mechanism to enable the C-glycosylation of proteins. These results afford opportunities for understanding human disease and therapeutic targeting of specific CMT paralogs.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
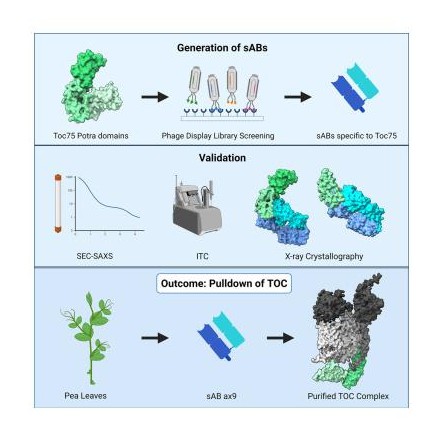
Srinivasan, Karthik; Erramilli, Satchal K; Chakravarthy, Srinivas; Gonzalez, Adrian; Kossiakoff, Anthony; Noinaj, Nicholas
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum Journal Article
In: Structure, vol. 31, no. 5, pp. 595–606.e5, 2023, ISSN: 1878-4186.
@article{pmid36977410,
title = {Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum},
author = {Karthik Srinivasan and Satchal K Erramilli and Srinivas Chakravarthy and Adrian Gonzalez and Anthony Kossiakoff and Nicholas Noinaj},
doi = {10.1016/j.str.2023.03.002},
issn = {1878-4186},
year = {2023},
date = {2023-05-01},
urldate = {2023-05-01},
journal = {Structure},
volume = {31},
number = {5},
pages = {595--606.e5},
abstract = {Roughly 95% of the proteins that make up the chloroplast must be imported from the cytoplasm. The machinery responsible for the translocation of these cargo proteins is called the translocon at the outer membrane of chloroplast (TOC). The TOC core consists of three proteins, Toc34, Toc75, and Toc159; no high-resolution structure has been solved of fully assembled TOC from plants. Efforts toward determining the structure of the TOC have been hindered almost entirely by difficulties in producing sufficient yields for structural studies. In this study, we introduce an innovative method that utilizes synthetic antigen binding fragments (sABs) to isolate TOC directly from wild-type plant biomass including A. thaliana and P. sativum. Binding between the sABs and the POTRA domains was characterized by size-exclusion chromatography coupled with small-angle X-ray scattering (SEC-SAXS), X-ray crystallography, and isothermal titration calorimetry. We also demonstrate the isolation of the TOC from P. sativum, laying the framework for large-scale isolation and purification of TOC for functional and structural studies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
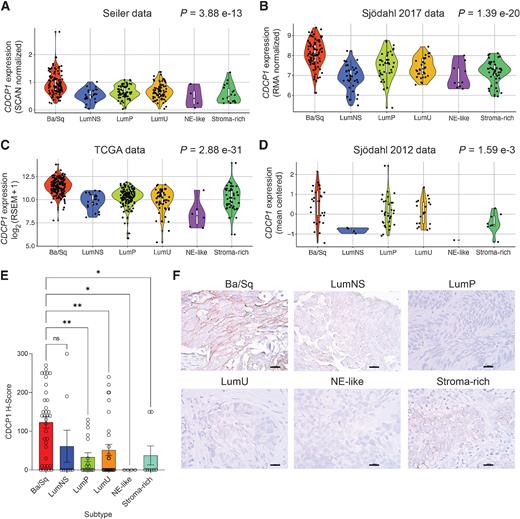
Chopra, Shalini; Trepka, Kai; Sakhamuri, Sasank; Carretero-González, Alberto; Zhu, Jun; Egusa, Emily; Zhou, Jie; Leung, Kevin; Zhao, Ning; Hooshdaran, Nima; Feng, Felix Y; Wells, James A; Chou, Jonathan; Evans, Michael J
Theranostic Targeting of CUB Domain-Containing Protein 1 (CDCP1) in Multiple Subtypes of Bladder Cancer Journal Article
In: Clin Cancer Res, vol. 29, no. 7, pp. 1232–1242, 2023, ISSN: 1557-3265.
@article{pmid36648492,
title = {Theranostic Targeting of CUB Domain-Containing Protein 1 (CDCP1) in Multiple Subtypes of Bladder Cancer},
author = {Shalini Chopra and Kai Trepka and Sasank Sakhamuri and Alberto Carretero-González and Jun Zhu and Emily Egusa and Jie Zhou and Kevin Leung and Ning Zhao and Nima Hooshdaran and Felix Y Feng and James A Wells and Jonathan Chou and Michael J Evans},
doi = {10.1158/1078-0432.CCR-22-1973},
issn = {1557-3265},
year = {2023},
date = {2023-04-01},
urldate = {2023-04-01},
journal = {Clin Cancer Res},
volume = {29},
number = {7},
pages = {1232--1242},
abstract = {PURPOSE: Despite recent approvals for checkpoint inhibitors and antibody-drug conjugates targeting NECTIN4 or TROP2, metastatic bladder cancer remains incurable and new treatment strategies are urgently needed. CUB domain-containing protein 1 (CDCP1) is a cell surface protein and promising drug target for many cancers. This study aimed to determine whether CDCP1 is expressed in bladder cancer and whether CDCP1 can be targeted for treatment with radiolabeled antibodies.nnEXPERIMENTAL DESIGN: CDCP1 expression was evaluated in four bladder cancer datasets (n = 1,047 biopsies). A tissue microarray of primary bladder cancer biopsies was probed for CDCP1 by IHC. CDCP1 expression was evaluated in patient-derived xenografts and cell lysates by immunoblot, flow cytometry, and saturation binding assays. Tumor detection in mouse bladder cancer models was tested using 89Zr-labeled 4A06, a monoclonal antibody targeting the ectodomain of CDCP1. 177Lu-4A06 was applied to mice bearing UMUC3 or HT-1376 xenografts to evaluate antitumor effects (CDCP1 expression in UMUC3 is 10-fold higher than HT-1376).nnRESULTS: CDCP1 was highest in the basal/squamous subtype, and CDCP1 was expressed in 53% of primary biopsies. CDCP1 was not correlated with pathologic or tumor stage, metastatic site, or NECTIN4 and TROP2 at the mRNA or protein level. CDCP1 ranged from 105 to 106 receptors per cell. Mechanism studies showed that RAS signaling induced CDCP1 expression. 89Zr-4A06 PET detected five human bladder cancer xenografts. 177Lu-4A06 inhibited the growth of UMUC3 and HT-1376 xenografts, models with high and moderate CDCP1 expression, respectively.nnCONCLUSIONS: These data establish that CDCP1 is expressed in bladder cancer, including TROP2 and NECTIN4-null disease, and suggest that bladder cancer can be treated with CDCP1-targeted radiotherapy.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Rettko, Nicholas J; Kirkemo, Lisa L; Wells, James A
Secreted HLA Fc-Fusion Profiles Immunopeptidome in Hypoxic PDAC and Cellular Senescence Journal Article
In: bioRxiv, 2023, ISSN: 2692-8205.
@article{pmid37090675,
title = {Secreted HLA Fc-Fusion Profiles Immunopeptidome in Hypoxic PDAC and Cellular Senescence},
author = {Nicholas J Rettko and Lisa L Kirkemo and James A Wells},
doi = {10.1101/2023.04.10.536290},
issn = {2692-8205},
year = {2023},
date = {2023-04-01},
urldate = {2023-04-01},
journal = {bioRxiv},
abstract = {Human leukocyte antigens (HLA) display peptides largely from intracellular proteins on the surface of cells in major histocompatibility complex (MHC)-peptide complexes. These complexes provide a biological window into the cell, and peptides derived from disease-associated antigens can serve as biomarkers and therapeutic targets. Thus, proper identification of peptides and the corresponding presenting HLA allele in disease phenotypes is important for the design and execution of therapeutic strategies using engineered T-cell receptors or antibodies. Yet, current mass spectrometry methods for profiling the immunopeptidome typically require large and complex sample inputs, complicating the study of several disease phenotypes and lowering the confidence of both peptide and allele identification. Here, we describe a novel secreted HLA (sHLA) Fc-fusion construct that allows for simple peptide identification from single HLA alleles in two important disease models: hypoxic pancreatic ductal adenocarcinoma (PDAC) and cellular senescence. We identify hypoxia and senescence-associated peptides that could act as future targets for immunotherapy. More generally, the method streamlines the time between sample preparation and injection from days to hours, yielding allele-restricted target identification in a temporally controlled manner. Overall, this method identified >30,000 unique HLA-associated peptides across two different HLA alleles and seven cell lines. Notably, ∼9,300 of these unique HLA-associated peptides had previously not been identified in the Immune Epitope Database. We believe the sHLA Fc-fusion capture technology will accelerate the study of the immunopeptidome as therapeutic interest in HLA-peptide complexes increases in cancer and beyond.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
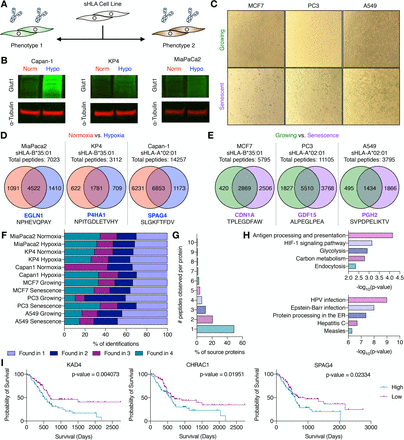
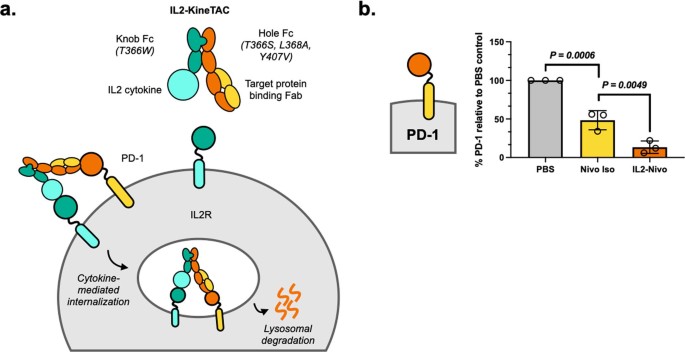
Pance, Katarina; Gramespacher, Josef A; Byrnes, James R; Salangsang, Fernando; Serrano, Juan-Antonio C; Cotton, Adam D; Steri, Veronica; Wells, James A
Modular cytokine receptor-targeting chimeras for targeted degradation of cell surface and extracellular proteins Journal Article
In: Nat Biotechnol, vol. 41, no. 2, pp. 273–281, 2023, ISSN: 1546-1696.
@article{pmid36138170,
title = {Modular cytokine receptor-targeting chimeras for targeted degradation of cell surface and extracellular proteins},
author = {Katarina Pance and Josef A Gramespacher and James R Byrnes and Fernando Salangsang and Juan-Antonio C Serrano and Adam D Cotton and Veronica Steri and James A Wells},
doi = {10.1038/s41587-022-01456-2},
issn = {1546-1696},
year = {2023},
date = {2023-02-01},
urldate = {2023-02-01},
journal = {Nat Biotechnol},
volume = {41},
number = {2},
pages = {273--281},
abstract = {Targeted degradation of cell surface and extracellular proteins via lysosomal delivery is an important means to modulate extracellular biology. However, these approaches have limitations due to lack of modularity, ease of development, restricted tissue targeting and applicability to both cell surface and extracellular proteins. We describe a lysosomal degradation strategy, termed cytokine receptor-targeting chimeras (KineTACs), that addresses these limitations. KineTACs are fully genetically encoded bispecific antibodies consisting of a cytokine arm, which binds its cognate cytokine receptor, and a target-binding arm for the protein of interest. We show that KineTACs containing the cytokine CXCL12 can use the decoy recycling receptor, CXCR7, to target a variety of target proteins to the lysosome for degradation. Additional KineTACs were designed to harness other CXCR7-targeting cytokines, CXCL11 and vMIPII, and the interleukin-2 (IL-2) receptor-targeting cytokine IL-2. Thus, KineTACs represent a general, modular, selective and simple genetically encoded strategy for inducing lysosomal delivery of extracellular and cell surface targets with broad or tissue-specific distribution.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Kordon, Szymon P; Dutka, Przemysław; Adamska, Justyna M; Bandekar, Sumit J; Leon, Katherine; Erramilli, Satchal K; Adams, Brock; Li, Jingxian; Kossiakoff, Anthony A; Araç, Demet
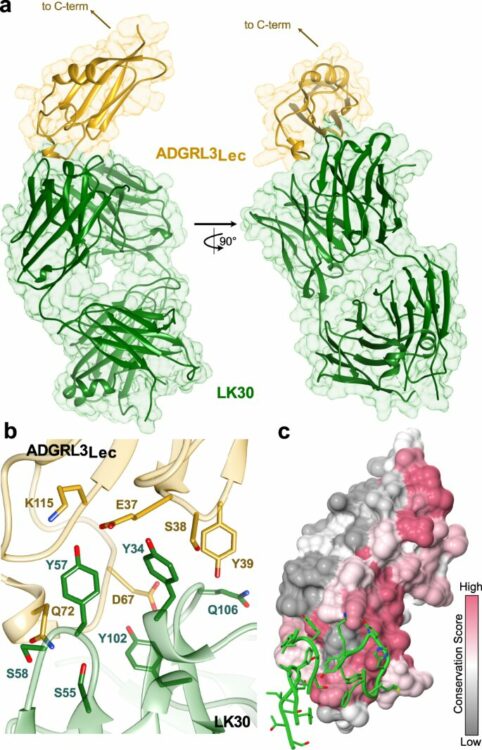
Isoform- and ligand-specific modulation of the adhesion GPCR ADGRL3/Latrophilin3 by a synthetic binder Journal Article
In: Nat Commun, vol. 14, no. 1, pp. 635, 2023, ISSN: 2041-1723.
@article{pmid36746957,
title = {Isoform- and ligand-specific modulation of the adhesion GPCR ADGRL3/Latrophilin3 by a synthetic binder},
author = {Szymon P Kordon and Przemysław Dutka and Justyna M Adamska and Sumit J Bandekar and Katherine Leon and Satchal K Erramilli and Brock Adams and Jingxian Li and Anthony A Kossiakoff and Demet Araç},
doi = {10.1038/s41467-023-36312-7},
issn = {2041-1723},
year = {2023},
date = {2023-02-01},
urldate = {2023-02-01},
journal = {Nat Commun},
volume = {14},
number = {1},
pages = {635},
abstract = {Adhesion G protein-coupled receptors (aGPCRs) are cell-surface proteins with large extracellular regions that bind to multiple ligands to regulate key biological functions including neurodevelopment and organogenesis. Modulating a single function of a specific aGPCR isoform while affecting no other function and no other receptor is not trivial. Here, we engineered an antibody, termed LK30, that binds to the extracellular region of the aGPCR ADGRL3, and specifically acts as an agonist for ADGRL3 but not for its isoform, ADGRL1. The LK30/ADGRL3 complex structure revealed that the LK30 binding site on ADGRL3 overlaps with the binding site for an ADGRL3 ligand - teneurin. In cellular-adhesion assays, LK30 specifically broke the trans-cellular interaction of ADGRL3 with teneurin, but not with another ADGRL3 ligand - FLRT3. Our work provides proof of concept for the modulation of isoform- and ligand-specific aGPCR functions using unique tools, and thus establishes a foundation for the development of fine-tuned aGPCR-targeted therapeutics.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}