Publications: 2022
2022
Cotton, Adam D; Wells, James A; Seiple, Ian B
Biotin as a Reactive Handle to Selectively Label Proteins and DNA with Small Molecules Journal Article
In: ACS Chem Biol, vol. 17, no. 12, pp. 3270–3275, 2022, ISSN: 1554-8937.
@article{pmid34410115,
title = {Biotin as a Reactive Handle to Selectively Label Proteins and DNA with Small Molecules},
author = {Adam D Cotton and James A Wells and Ian B Seiple},
doi = {10.1021/acschembio.1c00252},
issn = {1554-8937},
year = {2022},
date = {2022-12-01},
urldate = {2022-12-01},
journal = {ACS Chem Biol},
volume = {17},
number = {12},
pages = {3270--3275},
abstract = {Biotin is a common functional handle for bioconjugation to proteins and DNA, but its uses are limited to protein-containing conjugation partners such as streptavidin and derivatives thereof. Recently, oxaziridine reagents were developed that selectively conjugate the thioether of methionines on the surface of proteins, a method termed redox-activated chemical tagging (ReACT). These reagents generate sulfimide linkages that range in stability depending on the solvent accessibility and substitutions on the oxaziridine. Here we show that oxaziridine reagents react rapidly with the thioether in biotin to produce sulfimide products that are stable for more than 10 d at 37 °C. This method, which we call biotin redox-activated chemical tagging (BioReACT), expands the utility of biotin labeling and enables a predictable and stable chemical conjugation to biomolecules without the need to screen for a suitable methionine conjugation site. We demonstrate the versatility of this approach by producing a fluorescently labeled antibody, an antibody-drug conjugate, and a small molecule-conjugated oligonucleotide. We anticipate that BioReACT will be useful to rapidly introduce biorthogonal handles into biomolecules using biotin, a functional group that is widespread and straightforward to install.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Stopfer, Lauren E; Rettko, Nicholas J; Leddy, Owen; Mesfin, Joshua M; Brown, Eric; Winski, Shannon; Bryson, Bryan; Wells, James A; White, Forest M
MEK inhibition enhances presentation of targetable MHC-I tumor antigens in mutant melanomas Journal Article
In: Proc Natl Acad Sci U S A, vol. 119, no. 49, pp. e2208900119, 2022, ISSN: 1091-6490.
@article{pmid36454758,
title = {MEK inhibition enhances presentation of targetable MHC-I tumor antigens in mutant melanomas},
author = {Lauren E Stopfer and Nicholas J Rettko and Owen Leddy and Joshua M Mesfin and Eric Brown and Shannon Winski and Bryan Bryson and James A Wells and Forest M White},
doi = {10.1073/pnas.2208900119},
issn = {1091-6490},
year = {2022},
date = {2022-12-01},
urldate = {2022-12-01},
journal = {Proc Natl Acad Sci U S A},
volume = {119},
number = {49},
pages = {e2208900119},
abstract = {Combining multiple therapeutic strategies in NRAS/BRAF mutant melanoma-namely MEK/BRAF kinase inhibitors, immune checkpoint inhibitors (ICIs), and targeted immunotherapies-may offer an improved survival benefit by overcoming limitations associated with any individual therapy. Still, optimal combination, order, and timing of administration remains under investigation. Here, we measure how MEK inhibition (MEKi) alters anti-tumor immunity by utilizing quantitative immunopeptidomics to profile changes in the peptide major histocompatibility molecules (pMHC) repertoire. These data reveal a collection of tumor antigens whose presentation levels are selectively augmented following therapy, including several epitopes present at over 1,000 copies per cell. We leveraged the tunable abundance of MEKi-modulated antigens by targeting four epitopes with pMHC-specific T cell engagers and antibody drug conjugates, enhancing cell killing in tumor cells following MEK inhibition. These results highlight drug treatment as a means to enhance immunotherapy efficacy by targeting specific upregulated pMHCs and provide a methodological framework for identifying, quantifying, and therapeutically targeting additional epitopes of interest.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Elledge, Susanna K; Eigl, Ian; Phelps, Maira; McClinton, Khayla; Zhou, Xin X; Leung, Kevin K; Tato, Cristina M; Wells, James A
Using Split Luminescent Biosensors for SARS-CoV-2 Antibody Detection in Serum, Plasma, and Blood Samples Journal Article
In: Curr Protoc, vol. 2, no. 10, pp. e521, 2022, ISSN: 2691-1299.
@article{pmid36200787,
title = {Using Split Luminescent Biosensors for SARS-CoV-2 Antibody Detection in Serum, Plasma, and Blood Samples},
author = {Susanna K Elledge and Ian Eigl and Maira Phelps and Khayla McClinton and Xin X Zhou and Kevin K Leung and Cristina M Tato and James A Wells},
doi = {10.1002/cpz1.521},
issn = {2691-1299},
year = {2022},
date = {2022-10-01},
urldate = {2022-10-01},
journal = {Curr Protoc},
volume = {2},
number = {10},
pages = {e521},
abstract = {Antibody detection assays are essential for evaluating immunity of individuals against a given virus, and this has been particularly relevant during the COVID-19 pandemic. Current serology assays either require a laboratory setting and take >1 hr (i.e., enzyme-linked immunosorbent assay [ELISA]) or are rapid but only qualitative in nature and cannot accurately track antibody levels over time (i.e., lateral flow assay [LFA]). Therefore, there is a need for development of a rapid and simple but also quantitative assay that can evaluate antibody levels in patients accurately over time. We have developed an assay that uses a split nanoluciferase fused to the spike or nucleocapsid proteins of the SARS-CoV-2 virus to enable luminescent-based detection of spike- or nucleocapsid-binding antibodies in serum, plasma, and whole blood samples. The resulting approach is simple, rapid, and quantitative and is highly amenable to low-/medium-throughput scale using plate-based assays, high-throughput scale using robotics, and point-of-care applications. In this article, we describe how to perform the assay in a laboratory setting using a plate reader or liquid-handling robotics and in a point-of-care setting using a handheld, battery-powered luminometer. Together, these assays allow antibody detection to be easily performed in multiple settings by simplifying and reducing assay time in a laboratory or clinical environment and by allowing for antibody detection in point-of-care, nonlaboratory settings. © 2022 Wiley Periodicals LLC. Basic Protocol: SARS-CoV-2 antibody detection using the split-luciferase assay on a medium-throughput scale with a laboratory luminometer Alternate Protocol 1: High-throughput-based protocol for SARS-CoV-2 antibody detection using a robotic platform Alternate Protocol 2: Point-of-care-based protocol for SARS-CoV-2 antibody detection using a handheld luminometer Support Protocol: Determining positive/negative cutoffs for test samples and standardizing the assay between days.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Schaefer, Kaitlin; Lui, Irene; Byrnes, James R; Kang, Emily; Zhou, Jie; Weeks, Amy M; Wells, James A
Direct Identification of Proteolytic Cleavages on Living Cells Using a Glycan-Tethered Peptide Ligase Journal Article
In: ACS Cent Sci, vol. 8, no. 10, pp. 1447–1456, 2022, ISSN: 2374-7943.
@article{pmid36313159,
title = {Direct Identification of Proteolytic Cleavages on Living Cells Using a Glycan-Tethered Peptide Ligase},
author = {Kaitlin Schaefer and Irene Lui and James R Byrnes and Emily Kang and Jie Zhou and Amy M Weeks and James A Wells},
doi = {10.1021/acscentsci.2c00899},
issn = {2374-7943},
year = {2022},
date = {2022-10-01},
urldate = {2022-10-01},
journal = {ACS Cent Sci},
volume = {8},
number = {10},
pages = {1447--1456},
abstract = {Proteolytic cleavage of cell surface proteins triggers critical processes including cell-cell interactions, receptor activation, and shedding of signaling proteins. Consequently, dysregulated extracellular proteases contribute to malignant cell phenotypes including most cancers. To understand these effects, methods are needed that identify proteolyzed membrane proteins within diverse cellular contexts. Herein we report a proteomic approach, called cell surface N-terminomics, to broadly identify precise cleavage sites (neo-N-termini) on the surface of living cells. First, we functionalized the engineered peptide ligase, called stabiligase, with an N-terminal nucleophile that enables covalent attachment to naturally occurring glycans. Upon the addition of a biotinylated peptide ester, glycan-tethered stabiligase efficiently tags extracellular neo-N-termini for proteomic analysis. To demonstrate the versatility of this approach, we identified and characterized 1532 extracellular neo-N-termini across a panel of different cell types including primary immune cells. The vast majority of cleavages were not identified by previous proteomic studies. Lastly, we demonstrated that single oncogenes, and , induce extracellular proteolytic remodeling of proteins involved in cancerous cell growth, invasion, and migration. Cell surface N-terminomics is a generalizable platform that can reveal proteolyzed, neoepitopes to target using immunotherapies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
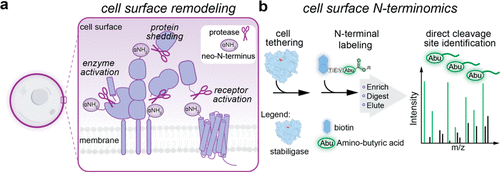
Orlando, Benjamin J; Dominik, Pawel K; Roy, Sourav; Ogbu, Chinemerem P; Erramilli, Satchal K; Kossiakoff, Anthony A; Vecchio, Alex J
Development, structure, and mechanism of synthetic antibodies that target claudin and Clostridium perfringens enterotoxin complexes Journal Article
In: J Biol Chem, vol. 298, no. 9, pp. 102357, 2022, ISSN: 1083-351X.
@article{pmid35952760,
title = {Development, structure, and mechanism of synthetic antibodies that target claudin and Clostridium perfringens enterotoxin complexes},
author = {Benjamin J Orlando and Pawel K Dominik and Sourav Roy and Chinemerem P Ogbu and Satchal K Erramilli and Anthony A Kossiakoff and Alex J Vecchio},
doi = {10.1016/j.jbc.2022.102357},
issn = {1083-351X},
year = {2022},
date = {2022-09-01},
urldate = {2022-09-01},
journal = {J Biol Chem},
volume = {298},
number = {9},
pages = {102357},
abstract = {Strains of Clostridium perfringens produce a two-domain enterotoxin (CpE) that afflicts humans and domesticated animals, causing prevalent gastrointestinal illnesses. CpE's C-terminal domain (cCpE) binds cell surface receptors, followed by a restructuring of its N-terminal domain to form a membrane-penetrating β-barrel pore, which is toxic to epithelial cells of the gut. The claudin family of membrane proteins are known receptors for CpE and also control the architecture and function of cell-cell contacts (tight junctions) that create barriers to intercellular molecular transport. CpE binding and assembly disables claudin barrier function and induces cytotoxicity via β-pore formation, disrupting gut homeostasis; however, a structural basis of this process and strategies to inhibit the claudin-CpE interactions that trigger it are both lacking. Here, we used a synthetic antigen-binding fragment (sFab) library to discover two sFabs that bind claudin-4 and cCpE complexes. We established these sFabs' mode of molecular recognition and binding properties and determined structures of each sFab bound to claudin-4-cCpE complexes using cryo-EM. The structures reveal that the sFabs bind a shared epitope, but conform distinctly, which explains their unique binding equilibria. Mutagenesis of antigen/sFab interfaces observed therein result in binding changes, validating the structures, and uncovering the sFab's targeting mechanism. From these insights, we generated a model for CpE's claudin-bound β-pore that predicted sFabs would not prevent cytotoxicity, which we then verified in vivo. Taken together, this work demonstrates the development and mechanism of claudin/cCpE-binding sFabs that provide a framework and strategy for obstructing claudin/CpE assembly to treat CpE-linked gastrointestinal diseases.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
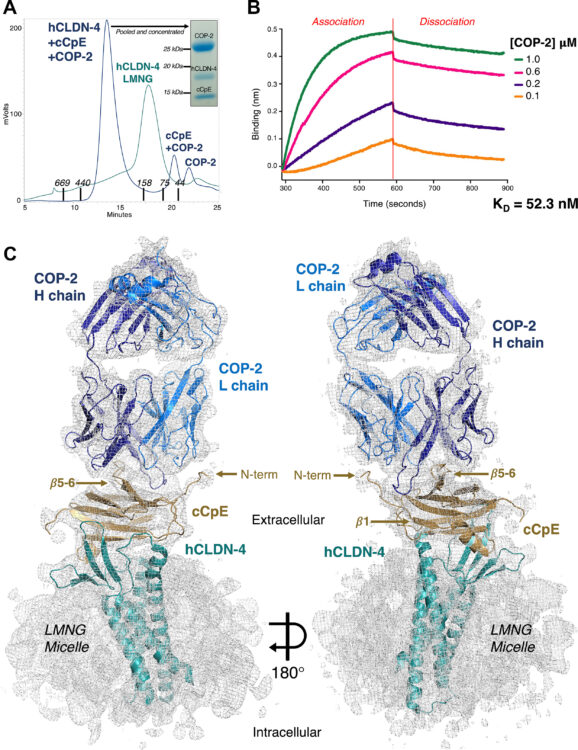
Remesh, Soumya G; Merz, Gregory E; Brilot, Axel F; Chio, Un Seng; Rizo, Alexandrea N; Pospiech, Thomas H; Lui, Irene; Laurie, Mathew T; Glasgow, Jeff; Le, Chau Q; Zhang, Yun; Diwanji, Devan; Hernandez, Evelyn; Lopez, Jocelyne; Pawar, Komal Ishwar; Pourmal, Sergei; Smith, Amber M; Zhou, Fengbo; and Joseph DeRisi,; Kortemme, Tanja; Rosenberg, Oren S; Glasgow, Anum; Leung, Kevin K; Wells, James A; Verba, Kliment A
Computational pipeline provides mechanistic understanding of Omicron variant of concern neutralizing engineered ACE2 receptor traps Journal Article
In: bioRxiv, 2022, ISSN: 2692-8205.
@article{pmid35982665,
title = {Computational pipeline provides mechanistic understanding of Omicron variant of concern neutralizing engineered ACE2 receptor traps},
author = {Soumya G Remesh and Gregory E Merz and Axel F Brilot and Un Seng Chio and Alexandrea N Rizo and Thomas H Pospiech and Irene Lui and Mathew T Laurie and Jeff Glasgow and Chau Q Le and Yun Zhang and Devan Diwanji and Evelyn Hernandez and Jocelyne Lopez and Komal Ishwar Pawar and Sergei Pourmal and Amber M Smith and Fengbo Zhou and and Joseph DeRisi and Tanja Kortemme and Oren S Rosenberg and Anum Glasgow and Kevin K Leung and James A Wells and Kliment A Verba},
doi = {10.1101/2022.08.09.503400},
issn = {2692-8205},
year = {2022},
date = {2022-08-01},
urldate = {2022-08-01},
journal = {bioRxiv},
abstract = {The SARS-CoV-2 Omicron variant, with 15 mutations in Spike receptor binding domain (Spike-RBD), renders virtually all clinical monoclonal antibodies against WT SARS-CoV-2 ineffective. We recently engineered the SARS-CoV-2 host entry receptor, ACE2, to tightly bind WT-Spike-RBD and prevent viral entry into host cells ("receptor traps"). Here we determine cryo-EM structures of our receptor traps in complex with full length Spike. We develop a multi-model pipeline combining Rosetta protein modeling software and cryo-EM to allow interface energy calculations even at limited resolution and identify interface side chains that allow for high affinity interactions between our ACE2 receptor traps and Spike-RBD. Our structural analysis provides a mechanistic rationale for the high affinity (0.53 - 4.2nM) binding of our ACE2 receptor traps to Omicron-RBD confirmed with biolayer interferometry measurements. Finally, we show that ACE2 receptor traps potently neutralize Omicron- and Delta-pseudotyped viruses, providing alternative therapeutic routes to combat this evolving virus.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Liu, Hongtao; Irobalieva, Rossitza N; Bang-Sørensen, Rose; Nosol, Kamil; Mukherjee, Somnath; Agrawal, Parth; Stieger, Bruno; Kossiakoff, Anthony A; Locher, Kaspar P
Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver Miscellaneous
2022, ISSN: 1748-7838.
@misc{pmid35726088,
title = {Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver},
author = {Hongtao Liu and Rossitza N Irobalieva and Rose Bang-Sørensen and Kamil Nosol and Somnath Mukherjee and Parth Agrawal and Bruno Stieger and Anthony A Kossiakoff and Kaspar P Locher},
doi = {10.1038/s41422-022-00680-4},
issn = {1748-7838},
year = {2022},
date = {2022-08-01},
urldate = {2022-08-01},
journal = {Cell Res},
volume = {32},
number = {8},
pages = {773--776},
keywords = {},
pubstate = {published},
tppubtype = {misc}
}
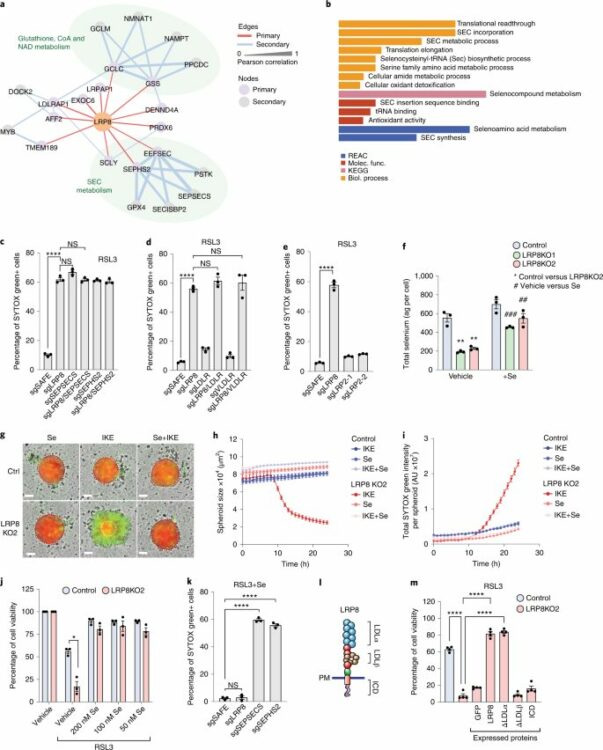
Li, Zhipeng; Ferguson, Lucas; Deol, Kirandeep K; Roberts, Melissa A; Magtanong, Leslie; Hendricks, Joseph M; Mousa, Gergey Alzaem; Kilinc, Seda; Schaefer, Kaitlin; Wells, James A; Bassik, Michael C; Goga, Andrei; Dixon, Scott J; Ingolia, Nicholas T; Olzmann, James A
Ribosome stalling during selenoprotein translation exposes a ferroptosis vulnerability Journal Article
In: Nat Chem Biol, vol. 18, no. 7, pp. 751–761, 2022, ISSN: 1552-4469.
@article{pmid35637349,
title = {Ribosome stalling during selenoprotein translation exposes a ferroptosis vulnerability},
author = {Zhipeng Li and Lucas Ferguson and Kirandeep K Deol and Melissa A Roberts and Leslie Magtanong and Joseph M Hendricks and Gergey Alzaem Mousa and Seda Kilinc and Kaitlin Schaefer and James A Wells and Michael C Bassik and Andrei Goga and Scott J Dixon and Nicholas T Ingolia and James A Olzmann},
doi = {10.1038/s41589-022-01033-3},
issn = {1552-4469},
year = {2022},
date = {2022-07-01},
urldate = {2022-07-01},
journal = {Nat Chem Biol},
volume = {18},
number = {7},
pages = {751--761},
abstract = {The selenoprotein glutathione peroxidase 4 (GPX4) prevents ferroptosis by converting lipid peroxides into nontoxic lipid alcohols. GPX4 has emerged as a promising therapeutic target for cancer treatment, but some cancer cells are resistant to ferroptosis triggered by GPX4 inhibition. Using a chemical-genetic screen, we identify LRP8 (also known as ApoER2) as a ferroptosis resistance factor that is upregulated in cancer. Loss of LRP8 decreases cellular selenium levels and the expression of a subset of selenoproteins. Counter to the canonical hierarchical selenoprotein regulatory program, GPX4 levels are strongly reduced due to impaired translation. Mechanistically, low selenium levels result in ribosome stalling at the inefficiently decoded GPX4 selenocysteine UGA codon, leading to ribosome collisions, early translation termination and proteasomal clearance of the N-terminal GPX4 fragment. These findings reveal rewiring of the selenoprotein hierarchy in cancer cells and identify ribosome stalling and collisions during GPX4 translation as ferroptosis vulnerabilities in cancer.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
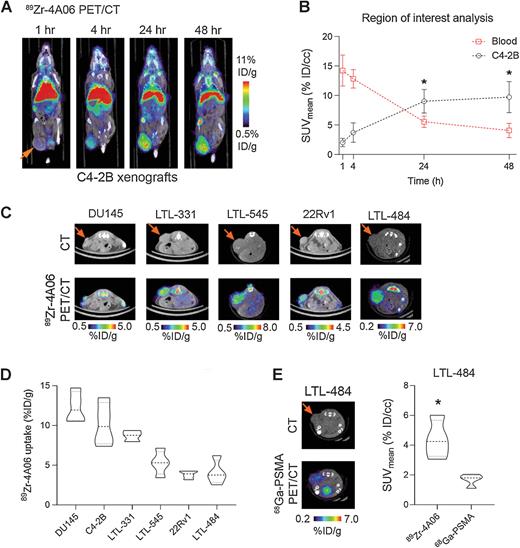
Zhao, Ning; Chopra, Shalini; Trepka, Kai; Wang, Yung-Hua; Sakhamuri, Sasank; Hooshdaran, Nima; Kim, Hyunjung; Zhou, Jie; Lim, Shion A; Leung, Kevin K; Egusa, Emily A; Zhu, Jun; Zhang, Li; Foye, Adam; Sriram, Renuka; Chan, Emily; Seo, Youngho; Feng, Felix Y; Small, Eric J; Chou, Jonathan; Wells, James A; Aggarwal, Rahul; Evans, Michael J
CUB Domain-Containing Protein 1 (CDCP1) Is a Target for Radioligand Therapy in Castration-Resistant Prostate Cancer, including PSMA Null Disease Journal Article
In: Clin Cancer Res, vol. 28, no. 14, pp. 3066–3075, 2022, ISSN: 1557-3265.
@article{pmid35604681,
title = {CUB Domain-Containing Protein 1 (CDCP1) Is a Target for Radioligand Therapy in Castration-Resistant Prostate Cancer, including PSMA Null Disease},
author = {Ning Zhao and Shalini Chopra and Kai Trepka and Yung-Hua Wang and Sasank Sakhamuri and Nima Hooshdaran and Hyunjung Kim and Jie Zhou and Shion A Lim and Kevin K Leung and Emily A Egusa and Jun Zhu and Li Zhang and Adam Foye and Renuka Sriram and Emily Chan and Youngho Seo and Felix Y Feng and Eric J Small and Jonathan Chou and James A Wells and Rahul Aggarwal and Michael J Evans},
doi = {10.1158/1078-0432.CCR-21-3858},
issn = {1557-3265},
year = {2022},
date = {2022-07-01},
urldate = {2022-07-01},
journal = {Clin Cancer Res},
volume = {28},
number = {14},
pages = {3066--3075},
abstract = {PURPOSE: With the improvement in overall survival with 177Lu-PSMA 617, radioligand therapy (RLT) is now a viable option for patients with metastatic castration-resistant prostate cancer (mCRPC). However, responses are variable, in part due to low PSMA expression in 30% of patients. Herein, we evaluated whether the cell surface protein CUB domain-containing protein 1 (CDCP1) can be exploited to treat mCRPC with RLT, including in PSMA-low subsets.nnEXPERIMENTAL DESIGN: CDCP1 levels were evaluated using RNA sequencing from 119 mCRPC biopsies. CDCP1 levels were assessed in 17 post-enzalutamide- or abiraterone-treated mCRPC biopsies, 12 patient-derived xenografts (PDX), and prostate cancer cell lines. 4A06, a recombinant human antibody that targets the CDCP1 ectodomain, was labeled with Zr-89 or Lu-177 and tested in tumor-bearing mice.nnRESULTS: CDCP1 expression was observed in 90% of mCRPC biopsies, including small-cell neuroendocrine (SCNC) and adenocarcinomas with low FOLH1 (PSMA) levels. Fifteen of 17 evaluable mCRPC biopsies (85%) demonstrated membranous CDCP1 expression, and 4 of 17 (23%) had higher CDCP1 H-scores compared with PSMA. CDCP1 was expressed in 10 of 12 PDX samples. Bmax values of approximately 22,000, 6,200, and 2,800 fmol/mg were calculated for PC3, DU145, and C4-2B human prostate cancer cells, respectively. 89Zr-4A06 PET detected six human prostate cancer xenografts, including PSMA-low tumors. 177Lu-4A06 significantly suppressed growth of DU145 and C4-2B xenografts.nnCONCLUSIONS: The data provide the first evidence supporting CDCP1-directed RLT to treat mCRPC. Expanded studies are warranted to determine whether CDCP1 is a viable drug target for patients with mCPRC.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
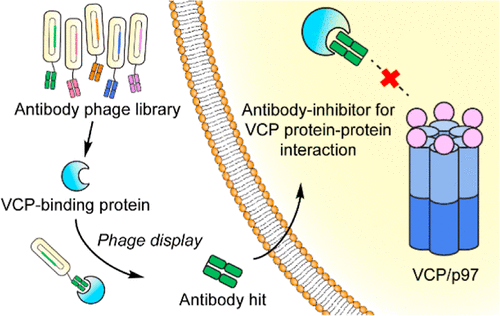
Jiang, Ziwen; Kuo, Yu-Hsuan; Zhong, Mengqi; Zhang, Jianchao; Zhou, Xin X; Xing, Lijuan; Wells, James A; Wang, Yanzhuang; Arkin, Michelle R
Adaptor-Specific Antibody Fragment Inhibitors for the Intracellular Modulation of p97 (VCP) Protein-Protein Interactions Journal Article
In: J Am Chem Soc, vol. 144, no. 29, pp. 13218–13225, 2022, ISSN: 1520-5126.
@article{pmid35819848,
title = {Adaptor-Specific Antibody Fragment Inhibitors for the Intracellular Modulation of p97 (VCP) Protein-Protein Interactions},
author = {Ziwen Jiang and Yu-Hsuan Kuo and Mengqi Zhong and Jianchao Zhang and Xin X Zhou and Lijuan Xing and James A Wells and Yanzhuang Wang and Michelle R Arkin},
doi = {10.1021/jacs.2c03665},
issn = {1520-5126},
year = {2022},
date = {2022-07-01},
urldate = {2022-07-01},
journal = {J Am Chem Soc},
volume = {144},
number = {29},
pages = {13218--13225},
abstract = {Protein-protein interactions (PPIs) form complex networks to drive cellular signaling and cellular functions. Precise modulation of a target PPI helps explain the role of the PPI in cellular events and possesses therapeutic potential. For example, valosin-containing protein (VCP/p97) is a hub protein that interacts with more than 30 adaptor proteins involved in various cellular functions. However, the role of each p97 PPI during the relevant cellular event is underexplored. The development of small-molecule PPI modulators remains challenging due to a lack of grooves and pockets in the relatively large PPI interface and the fact that a common binding groove in p97 binds to multiple adaptors. Here, we report an antibody fragment-based modulator for the PPI between p97 and its adaptor protein NSFL1C (p47). We engineered these antibody modulators by phage display against the p97-interacting domain of p47 and minimizing binding to other p97 adaptors. The selected antibody fragment modulators specifically disrupt the intracellular p97/p47 interaction. The potential of this antibody platform to develop PPI inhibitors in therapeutic applications was demonstrated through the inhibition of Golgi reassembly, which requires the p97/p47 interaction. This study presents a unique approach to modulate specific intracellular PPIs using engineered antibody fragments, demonstrating a method to dissect the function of a PPI within a convoluted PPI network.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
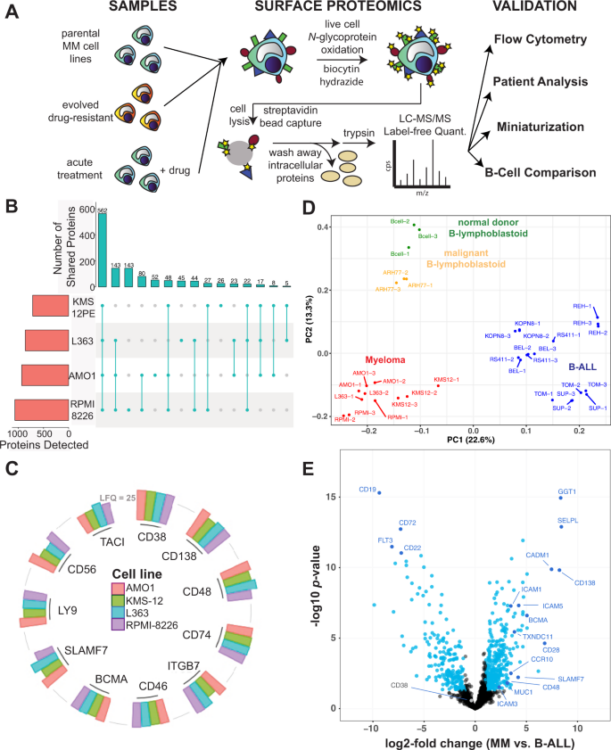
Ferguson, Ian D; Patiño-Escobar, Bonell; Tuomivaara, Sami T; Lin, Yu-Hsiu T; Nix, Matthew A; Leung, Kevin K; Kasap, Corynn; Ramos, Emilio; Vasquez, Wilson Nieves; Talbot, Alexis; Hale, Martina; Naik, Akul; Kishishita, Audrey; Choudhry, Priya; Lopez-Girona, Antonia; Miao, Weili; Wong, Sandy W; Wolf, Jeffrey L; Martin, Thomas G; Shah, Nina; Vandenberg, Scott; Prakash, Sonam; Besse, Lenka; Driessen, Christoph; Posey, Avery D; Mullins, R Dyche; Eyquem, Justin; Wells, James A; Wiita, Arun P
The surfaceome of multiple myeloma cells suggests potential immunotherapeutic strategies and protein markers of drug resistance Journal Article
In: Nat Commun, vol. 13, no. 1, pp. 4121, 2022, ISSN: 2041-1723.
@article{pmid35840578,
title = {The surfaceome of multiple myeloma cells suggests potential immunotherapeutic strategies and protein markers of drug resistance},
author = {Ian D Ferguson and Bonell Patiño-Escobar and Sami T Tuomivaara and Yu-Hsiu T Lin and Matthew A Nix and Kevin K Leung and Corynn Kasap and Emilio Ramos and Wilson Nieves Vasquez and Alexis Talbot and Martina Hale and Akul Naik and Audrey Kishishita and Priya Choudhry and Antonia Lopez-Girona and Weili Miao and Sandy W Wong and Jeffrey L Wolf and Thomas G Martin and Nina Shah and Scott Vandenberg and Sonam Prakash and Lenka Besse and Christoph Driessen and Avery D Posey and R Dyche Mullins and Justin Eyquem and James A Wells and Arun P Wiita},
doi = {10.1038/s41467-022-31810-6},
issn = {2041-1723},
year = {2022},
date = {2022-07-01},
urldate = {2022-07-01},
journal = {Nat Commun},
volume = {13},
number = {1},
pages = {4121},
abstract = {The myeloma surface proteome (surfaceome) determines tumor interaction with the microenvironment and serves as an emerging arena for therapeutic development. Here, we use glycoprotein capture proteomics to define the myeloma surfaceome at baseline, in drug resistance, and in response to acute drug treatment. We provide a scoring system for surface antigens and identify CCR10 as a promising target in this disease expressed widely on malignant plasma cells. We engineer proof-of-principle chimeric antigen receptor (CAR) T-cells targeting CCR10 using its natural ligand CCL27. In myeloma models we identify proteins that could serve as markers of resistance to bortezomib and lenalidomide, including CD53, CD10, EVI2B, and CD33. We find that acute lenalidomide treatment increases activity of MUC1-targeting CAR-T cells through antigen upregulation. Finally, we develop a miniaturized surface proteomic protocol for profiling primary plasma cell samples with low inputs. These approaches and datasets may contribute to the biological, therapeutic, and diagnostic understanding of myeloma.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
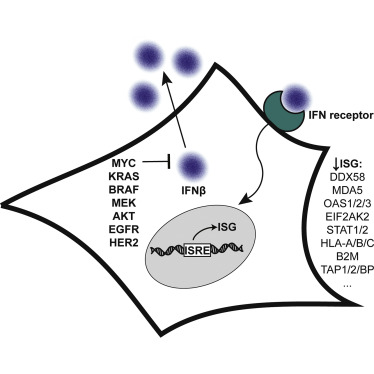
Solomon, Paige E; Kirkemo, Lisa L; Wilson, Gary M; Leung, Kevin K; Almond, Mark H; Sayles, Leanne C; Sweet-Cordero, E Alejandro; Rosenberg, Oren S; Coon, Joshua J; Wells, James A
In: Mol Cell Proteomics, vol. 21, no. 7, pp. 100247, 2022, ISSN: 1535-9484.
@article{pmid35594991,
title = {Discovery Proteomics Analysis Determines That Driver Oncogenes Suppress Antiviral Defense Pathways Through Reduction in Interferon-β Autocrine Stimulation},
author = {Paige E Solomon and Lisa L Kirkemo and Gary M Wilson and Kevin K Leung and Mark H Almond and Leanne C Sayles and E Alejandro Sweet-Cordero and Oren S Rosenberg and Joshua J Coon and James A Wells},
doi = {10.1016/j.mcpro.2022.100247},
issn = {1535-9484},
year = {2022},
date = {2022-07-01},
urldate = {2022-07-01},
journal = {Mol Cell Proteomics},
volume = {21},
number = {7},
pages = {100247},
abstract = {Since the discovery of oncogenes, there has been tremendous interest to understand their mechanistic basis and to develop broadly actionable therapeutics. Some of the most frequently activated oncogenes driving diverse cancers are c-MYC, EGFR, HER2, AKT, KRAS, BRAF, and MEK. Using a reductionist approach, we explored how cellular proteomes are remodeled in isogenic cell lines engineered with or without these driver oncogenes. The most striking discovery for all oncogenic models was the systematic downregulation of scores of antiviral proteins regulated by type 1 interferon. These findings extended to cancer cell lines and patient-derived xenograft models of highly refractory pancreatic cancer and osteosarcoma driven by KRAS and MYC oncogenes. The oncogenes reduced basal expression of and autocrine stimulation by type 1 interferon causing remarkable convergence on common phenotypic and functional profiles. In particular, there was dramatically lower expression of dsRNA sensors including DDX58 (RIG-I) and OAS proteins, which resulted in attenuated functional responses when the oncogenic cells were treated with the dsRNA mimetic, polyI:C, and increased susceptibility to infection with an RNA virus shown using SARS-CoV-2. Our reductionist approach provides molecular and functional insights connected to immune evasion hallmarks in cancers and suggests therapeutic opportunities.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
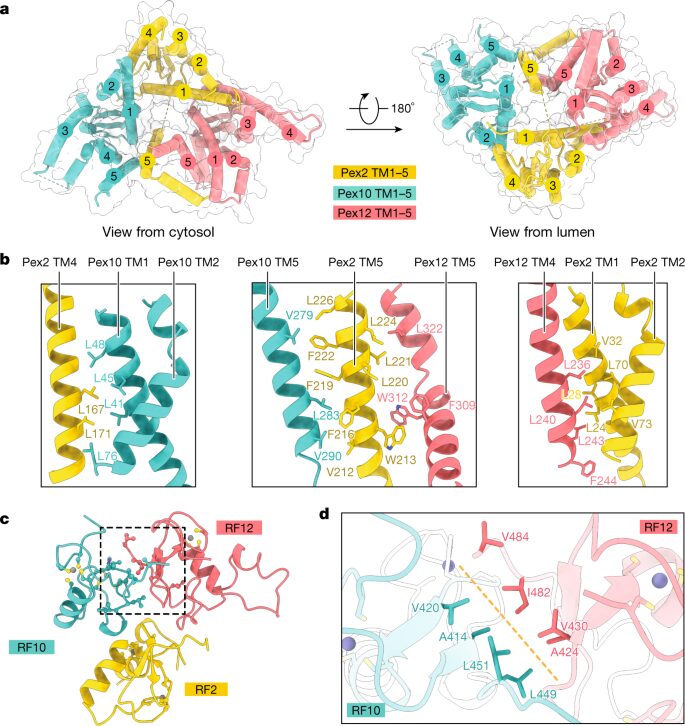
Feng, Peiqiang; Wu, Xudong; Erramilli, Satchal K; Paulo, Joao A; Knejski, Pawel; Gygi, Steven P; Kossiakoff, Anthony A; Rapoport, Tom A
A peroxisomal ubiquitin ligase complex forms a retrotranslocation channel Journal Article
In: Nature, vol. 607, no. 7918, pp. 374–380, 2022, ISSN: 1476-4687.
@article{pmid35768507,
title = {A peroxisomal ubiquitin ligase complex forms a retrotranslocation channel},
author = {Peiqiang Feng and Xudong Wu and Satchal K Erramilli and Joao A Paulo and Pawel Knejski and Steven P Gygi and Anthony A Kossiakoff and Tom A Rapoport},
doi = {10.1038/s41586-022-04903-x},
issn = {1476-4687},
year = {2022},
date = {2022-07-01},
urldate = {2022-07-01},
journal = {Nature},
volume = {607},
number = {7918},
pages = {374--380},
abstract = {Peroxisomes are ubiquitous organelles that house various metabolic reactions and are essential for human health. Luminal peroxisomal proteins are imported from the cytosol by mobile receptors, which then recycle back to the cytosol by a poorly understood process. Recycling requires receptor modification by a membrane-embedded ubiquitin ligase complex comprising three RING finger domain-containing proteins (Pex2, Pex10 and Pex12). Here we report a cryo-electron microscopy structure of the ligase complex, which together with biochemical and in vivo experiments reveals its function as a retrotranslocation channel for peroxisomal import receptors. Each subunit of the complex contributes five transmembrane segments that co-assemble into an open channel. The three ring finger domains form a cytosolic tower, with ring finger 2 (RF2) positioned above the channel pore. We propose that the N terminus of a recycling receptor is inserted from the peroxisomal lumen into the pore and monoubiquitylated by RF2 to enable extraction into the cytosol. If recycling is compromised, receptors are polyubiquitylated by the concerted action of RF10 and RF12 and degraded. This polyubiquitylation pathway also maintains the homeostasis of other peroxisomal import factors. Our results clarify a crucial step during peroxisomal protein import and reveal why mutations in the ligase complex cause human disease.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
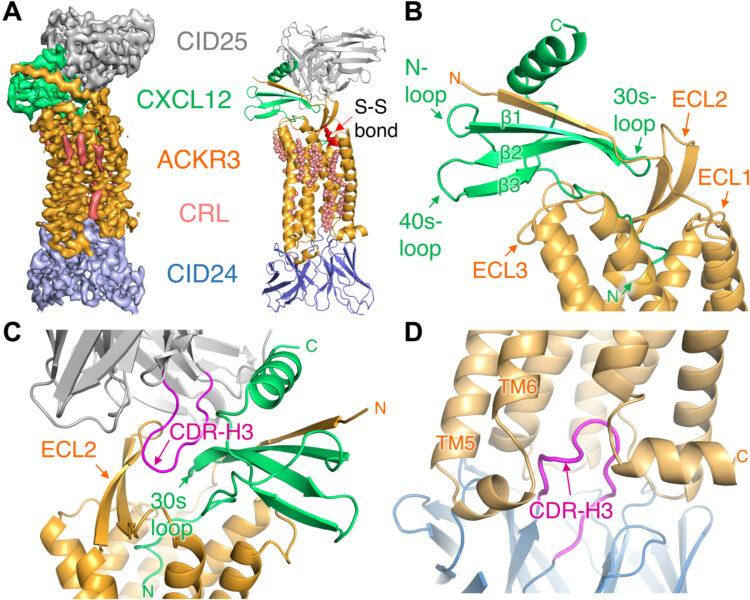
Yen, Yu-Chen; Schafer, Christopher T; Gustavsson, Martin; Eberle, Stefanie A; Dominik, Pawel K; Deneka, Dawid; Zhang, Penglie; Schall, Thomas J; Kossiakoff, Anthony A; Tesmer, John J G; Handel, Tracy M
Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias Journal Article
In: Sci Adv, vol. 8, no. 28, pp. eabn8063, 2022, ISSN: 2375-2548.
@article{pmid35857509,
title = {Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias},
author = {Yu-Chen Yen and Christopher T Schafer and Martin Gustavsson and Stefanie A Eberle and Pawel K Dominik and Dawid Deneka and Penglie Zhang and Thomas J Schall and Anthony A Kossiakoff and John J G Tesmer and Tracy M Handel},
doi = {10.1126/sciadv.abn8063},
issn = {2375-2548},
year = {2022},
date = {2022-07-01},
urldate = {2022-07-01},
journal = {Sci Adv},
volume = {8},
number = {28},
pages = {eabn8063},
abstract = {Both CXC chemokine receptor 4 (CXCR4) and atypical chemokine receptor 3 (ACKR3) are activated by the chemokine CXCL12 yet evoke distinct cellular responses. CXCR4 is a canonical G protein-coupled receptor (GPCR), whereas ACKR3 is intrinsically biased for arrestin. The molecular basis for this difference is not understood. Here, we describe cryo-EM structures of ACKR3 in complex with CXCL12, a more potent CXCL12 variant, and a small-molecule agonist. The bound chemokines adopt an unexpected pose relative to those established for CXCR4 and observed in other receptor-chemokine complexes. Along with functional studies, these structures provide insight into the ligand-binding promiscuity of ACKR3, why it fails to couple to G proteins, and its bias toward β-arrestin. The results lay the groundwork for understanding the physiological interplay of ACKR3 with other GPCRs.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
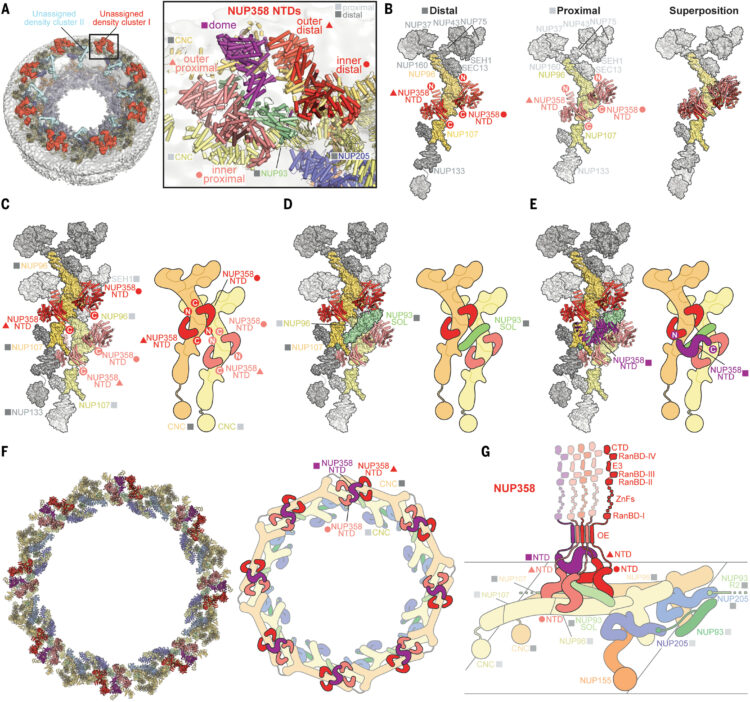
Bley, Christopher J; Nie, Si; Mobbs, George W; Petrovic, Stefan; Gres, Anna T; Liu, Xiaoyu; Mukherjee, Somnath; Harvey, Sho; Huber, Ferdinand M; Lin, Daniel H; Brown, Bonnie; Tang, Aaron W; Rundlet, Emily J; Correia, Ana R; Chen, Shane; Regmi, Saroj G; Stevens, Taylor A; Jette, Claudia A; Dasso, Mary; Patke, Alina; Palazzo, Alexander F; Kossiakoff, Anthony A; Hoelz, André
Architecture of the cytoplasmic face of the nuclear pore Journal Article
In: Science, vol. 376, no. 6598, pp. eabm9129, 2022, ISSN: 1095-9203.
@article{pmid35679405,
title = {Architecture of the cytoplasmic face of the nuclear pore},
author = {Christopher J Bley and Si Nie and George W Mobbs and Stefan Petrovic and Anna T Gres and Xiaoyu Liu and Somnath Mukherjee and Sho Harvey and Ferdinand M Huber and Daniel H Lin and Bonnie Brown and Aaron W Tang and Emily J Rundlet and Ana R Correia and Shane Chen and Saroj G Regmi and Taylor A Stevens and Claudia A Jette and Mary Dasso and Alina Patke and Alexander F Palazzo and Anthony A Kossiakoff and André Hoelz},
doi = {10.1126/science.abm9129},
issn = {1095-9203},
year = {2022},
date = {2022-06-01},
urldate = {2022-06-01},
journal = {Science},
volume = {376},
number = {6598},
pages = {eabm9129},
abstract = {INTRODUCTION The subcellular compartmentalization of eukaryotic cells requires selective transport of folded proteins and protein-nucleic acid complexes. Embedded in nuclear envelope pores, which are generated by the circumscribed fusion of the inner and outer nuclear membranes, nuclear pore complexes (NPCs) are the sole bidirectional gateways for nucleocytoplasmic transport. The ~110-MDa human NPC is an ~1000-protein assembly that comprises multiple copies of ~34 different proteins, collectively termed nucleoporins. The symmetric core of the NPC is composed of an inner ring encircling the central transport channel and outer rings formed by Y‑shaped coat nucleoporin complexes (CNCs) anchored atop both sides of the nuclear envelope. The outer rings are decorated with compartment‑specific asymmetric nuclear basket and cytoplasmic filament nucleoporins, which establish transport directionality and provide docking sites for transport factors and the small guanosine triphosphatase Ran. The cytoplasmic filament nucleoporins also play an essential role in the irreversible remodeling of messenger ribonucleoprotein particles (mRNPs) as they exit the central transport channel. Unsurprisingly, the NPC's cytoplasmic face represents a hotspot for disease‑associated mutations and is commonly targeted by viral virulence factors. RATIONALE Previous studies established a near-atomic composite structure of the human NPC's symmetric core by combining (i) biochemical reconstitution to elucidate the interaction network between symmetric nucleoporins, (ii) crystal and single-particle cryo-electron microscopy structure determination of nucleoporins and nucleoporin complexes to reveal their three-dimensional shape and the molecular details of their interactions, (iii) quantitative docking in cryo-electron tomography (cryo-ET) maps of the intact human NPC to uncover nucleoporin stoichiometry and positioning, and (iv) cell‑based assays to validate the physiological relevance of the biochemical and structural findings. In this work, we extended our approach to the cytoplasmic filament nucleoporins to reveal the near-atomic architecture of the cytoplasmic face of the human NPC. RESULTS Using biochemical reconstitution, we elucidated the protein-protein and protein-RNA interaction networks of the human and cytoplasmic filament nucleoporins, establishing an evolutionarily conserved heterohexameric cytoplasmic filament nucleoporin complex (CFNC) held together by a central heterotrimeric coiled‑coil hub that tethers two separate mRNP‑remodeling complexes. Further biochemical analysis and determination of a series of crystal structures revealed that the metazoan‑specific cytoplasmic filament nucleoporin NUP358 is composed of 16 distinct domains, including an N‑terminal S‑shaped α‑helical solenoid followed by a coiled‑coil oligomerization element, numerous Ran‑interacting domains, an E3 ligase domain, and a C‑terminal prolyl‑isomerase domain. Physiologically validated quantitative docking into cryo-ET maps of the intact human NPC revealed that pentameric NUP358 bundles, conjoined by the oligomerization element, are anchored through their N‑terminal domains to the central stalk regions of the CNC, projecting flexibly attached domains as far as ~600 Å into the cytoplasm. Using cell‑based assays, we demonstrated that NUP358 is dispensable for the architectural integrity of the assembled interphase NPC and RNA export but is required for efficient translation. After NUP358 assignment, the remaining 4-shaped cryo‑ET density matched the dimensions of the CFNC coiled‑coil hub, in close proximity to an outer-ring NUP93. Whereas the N-terminal NUP93 assembly sensor motif anchors the properly assembled related coiled‑coil channel nucleoporin heterotrimer to the inner ring, biochemical reconstitution confirmed that the NUP93 assembly sensor is reused in anchoring the CFNC to the cytoplasmic face of the human NPC. By contrast, two CFNCs are anchored by a divergent mechanism that involves assembly sensors located in unstructured portions of two CNC nucleoporins. Whereas unassigned cryo‑ET density occupies the NUP358 and CFNC binding sites on the nuclear face, docking of the nuclear basket component ELYS established that the equivalent position on the cytoplasmic face is unoccupied, suggesting that mechanisms other than steric competition promote asymmetric distribution of nucleoporins. CONCLUSION We have substantially advanced the biochemical and structural characterization of the asymmetric nucleoporins' architecture and attachment at the cytoplasmic and nuclear faces of the NPC. Our near‑atomic composite structure of the human NPC's cytoplasmic face provides a biochemical and structural framework for elucidating the molecular basis of mRNP remodeling, viral virulence factor interference with NPC function, and the underlying mechanisms of nucleoporin diseases at the cytoplasmic face of the NPC. [Figure: see text].},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
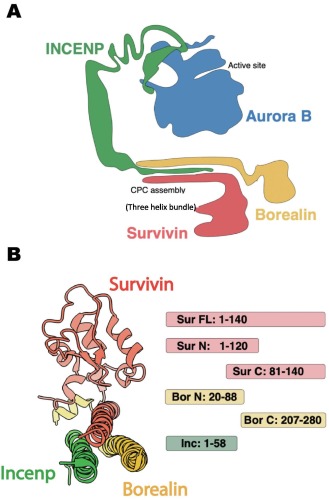
Ura, Marcin; Mukherjee, Somnath; Marcon, Edyta; Koestler, Stefan A; Kossiakoff, Anthony A
Synthetic Antibodies Detect Distinct Cellular States of Chromosome Passenger Complex Proteins Journal Article
In: J Mol Biol, vol. 434, no. 12, pp. 167602, 2022, ISSN: 1089-8638.
@article{pmid35469831,
title = {Synthetic Antibodies Detect Distinct Cellular States of Chromosome Passenger Complex Proteins},
author = {Marcin Ura and Somnath Mukherjee and Edyta Marcon and Stefan A Koestler and Anthony A Kossiakoff},
doi = {10.1016/j.jmb.2022.167602},
issn = {1089-8638},
year = {2022},
date = {2022-06-01},
urldate = {2022-06-01},
journal = {J Mol Biol},
volume = {434},
number = {12},
pages = {167602},
abstract = {High performance affinity reagents are essential tools to enable biologists to profile the cellular location and composition of macromolecular complexes undergoing dynamic reorganization. To support further development of such tools, we have assembled a high-throughput phage display pipeline to generate Fab-based affinity reagents that target different dynamic forms of a large macromolecular complex, using the Chromosomal Passenger Complex (CPC), as an example. The CPC is critical for the maintenance of chromosomal and cytoskeleton processes during cell division. The complex contains 4 protein components: Aurora B kinase, survivin, borealin and INCENP. The CPC acts as a node to dynamically organize other partnering subcomplexes to build multiple functional structures during mitotic progression. Using phage display mutagenesis, a cohort of synthetic antibodies (sABs) were generated against different domains of survivin, borealin and INCENP. Immunofluorescence established that a set of these sABs can discriminate between the form of the CPC complex in the midbody versus the spindle. Others localize to targets, which appear to be less organized, in the nucleus or cytoplasm. This differentiation suggests that different CPC epitopes have dynamic accessibility depending upon the mitotic state of the cell. An Immunoprecipitation/Mass Spectrometry analysis was performed using sABs that bound specifically to the CPC in either the midbody or MT spindle macromolecular assemblies. Thus, sABs can be exploited as high performance reagents to profile the accessibility of different components of the CPC within macromolecular assemblies during different stages of mitosis suggesting this high throughput approach will be applicable to other complex macromolecular systems.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
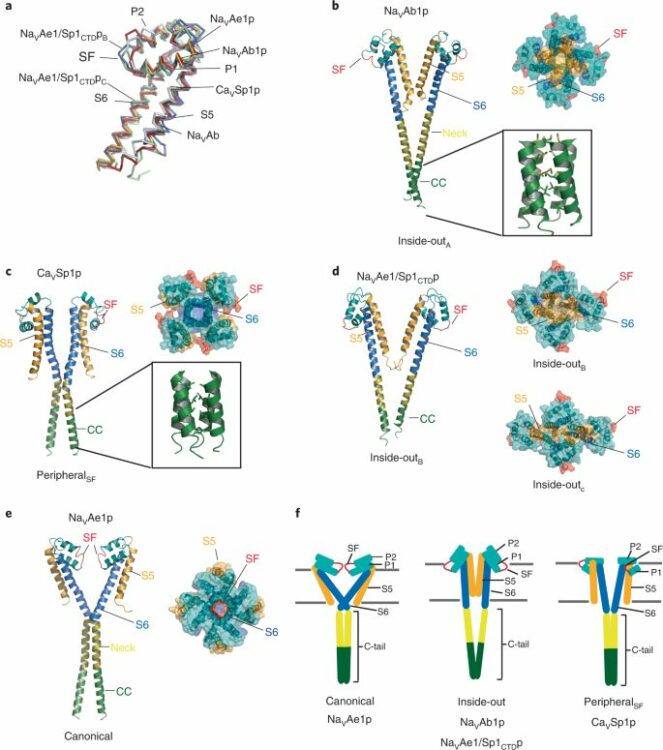
Arrigoni, Cristina; Lolicato, Marco; Shaya, David; Rohaim, Ahmed; Findeisen, Felix; Fong, Lam-Kiu; Colleran, Claire M; Dominik, Pawel; Kim, Sangwoo S; Schuermann, Jonathan P; DeGrado, William F; Grabe, Michael; Kossiakoff, Anthony A; Minor, Daniel L
Quaternary structure independent folding of voltage-gated ion channel pore domain subunits Journal Article
In: Nat Struct Mol Biol, vol. 29, no. 6, pp. 537–548, 2022, ISSN: 1545-9985.
@article{pmid35655098,
title = {Quaternary structure independent folding of voltage-gated ion channel pore domain subunits},
author = {Cristina Arrigoni and Marco Lolicato and David Shaya and Ahmed Rohaim and Felix Findeisen and Lam-Kiu Fong and Claire M Colleran and Pawel Dominik and Sangwoo S Kim and Jonathan P Schuermann and William F DeGrado and Michael Grabe and Anthony A Kossiakoff and Daniel L Minor},
doi = {10.1038/s41594-022-00775-x},
issn = {1545-9985},
year = {2022},
date = {2022-06-01},
urldate = {2022-06-01},
journal = {Nat Struct Mol Biol},
volume = {29},
number = {6},
pages = {537--548},
abstract = {Every voltage-gated ion channel (VGIC) has a pore domain (PD) made from four subunits, each comprising an antiparallel transmembrane helix pair bridged by a loop. The extent to which PD subunit structure requires quaternary interactions is unclear. Here, we present crystal structures of a set of bacterial voltage-gated sodium channel (BacNa) 'pore only' proteins that reveal a surprising collection of non-canonical quaternary arrangements in which the PD tertiary structure is maintained. This context-independent structural robustness, supported by molecular dynamics simulations, indicates that VGIC-PD tertiary structure is independent of quaternary interactions. This fold occurs throughout the VGIC superfamily and in diverse transmembrane and soluble proteins. Strikingly, characterization of PD subunit-binding Fabs indicates that non-canonical quaternary PD conformations can occur in full-length VGICs. Together, our data demonstrate that the VGIC-PD is an autonomously folded unit. This property has implications for VGIC biogenesis, understanding functional states, de novo channel design, and VGIC structural origins.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
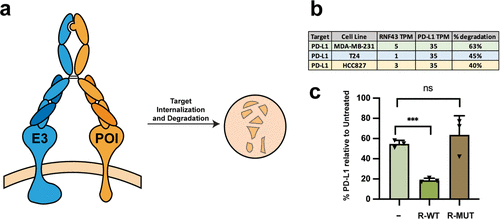
Gramespacher, Josef A; Cotton, Adam D; Burroughs, Paul W W; Seiple, Ian B; Wells, James A
Roadmap for Optimizing and Broadening Antibody-Based PROTACs for Degradation of Cell Surface Proteins Journal Article
In: ACS Chem Biol, vol. 17, no. 5, pp. 1259–1268, 2022, ISSN: 1554-8937.
@article{pmid35481357,
title = {Roadmap for Optimizing and Broadening Antibody-Based PROTACs for Degradation of Cell Surface Proteins},
author = {Josef A Gramespacher and Adam D Cotton and Paul W W Burroughs and Ian B Seiple and James A Wells},
doi = {10.1021/acschembio.2c00185},
issn = {1554-8937},
year = {2022},
date = {2022-05-01},
urldate = {2022-05-01},
journal = {ACS Chem Biol},
volume = {17},
number = {5},
pages = {1259--1268},
abstract = {Targeted protein degradation is a promising therapeutic strategy capable of overcoming the limitations of traditional occupancy-based inhibitors. By ablating all of the associated functions of a protein at once, the event-driven pharmacology of degrader technologies has recently enabled the targeting of proteins that have been historically deemed "undruggable". Most degradation strategies utilize the ubiquitin-proteasome system to mediate intracellular target degradation and are thus limited to targeting proteins with cytoplasmic domains. While some of these strategies, such as PROTACs, have shown great promise, there is a need for new modalities that can be applied to specifically target cell surface proteins. We previously described the development of an antibody-based PROTAC (AbTAC) that utilizes genetically encoded IgG bispecific antibody scaffolds to bring the cell surface E3-ligase RNF43 into the proximity of a membrane protein of interest (POI) to mediate its degradation. Here, we employ rational protein engineering strategies to interrogate and optimize the properties necessary for efficient degradation of two therapeutically important membrane proteins, PD-L1 and EGFR. We develop multiple antibodies to RNF43 and show that the specific antibody binding epitopes on RNF43 and the POI are more important than the affinities of the AbTAC antibodies. We further expand the available repertoire of E3 ligases by co-opting the E3-ligase ZNRF3 to degrade both PD-L1 and EGFR and show similar importance of epitope for degradation efficiency. Importantly, we show that both RNF43 and ZNRF3 AbTACs do not potentiate unwanted WNT signaling. Lastly, we find that these AbTACs can be even further improved by exploring various dual-binding and IgG scaffolds that range in flexibility, valency, and orientation of the binding arms. These structure-activity and mechanistic studies provide a roadmap for optimizing the development of AbTACs, thereby greatly expanding their utility for targeted cell surface protein degradation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
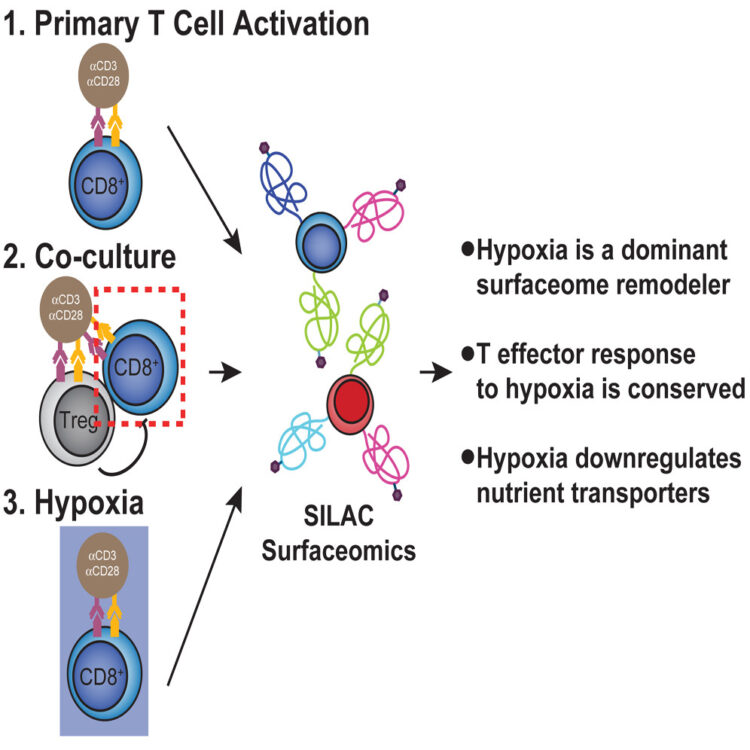
Byrnes, James R; Weeks, Amy M; Shifrut, Eric; Carnevale, Julia; Kirkemo, Lisa; Ashworth, Alan; Marson, Alexander; Wells, James A
Hypoxia Is a Dominant Remodeler of the Effector T Cell Surface Proteome Relative to Activation and Regulatory T Cell Suppression Journal Article
In: Mol Cell Proteomics, vol. 21, no. 4, pp. 100217, 2022, ISSN: 1535-9484.
@article{pmid35217172,
title = {Hypoxia Is a Dominant Remodeler of the Effector T Cell Surface Proteome Relative to Activation and Regulatory T Cell Suppression},
author = {James R Byrnes and Amy M Weeks and Eric Shifrut and Julia Carnevale and Lisa Kirkemo and Alan Ashworth and Alexander Marson and James A Wells},
doi = {10.1016/j.mcpro.2022.100217},
issn = {1535-9484},
year = {2022},
date = {2022-04-01},
urldate = {2022-04-01},
journal = {Mol Cell Proteomics},
volume = {21},
number = {4},
pages = {100217},
abstract = {Immunosuppressive factors in the tumor microenvironment (TME) impair T cell function and limit the antitumor immune response. T cell surface receptors and surface proteins that influence interactions and function in the TME are proven targets for cancer immunotherapy. However, how the entire surface proteome remodels in primary human T cells in response to specific suppressive factors in the TME remains to be broadly and systematically characterized. Here, using a reductionist cell culture approach with primary human T cells and stable isotopic labeling with amino acids in cell culture-based quantitative cell surface capture glycoproteomics, we examined how two immunosuppressive TME factors, regulatory T cells (Tregs) and hypoxia, globally affect the activated CD8 surface proteome (surfaceome). Surprisingly, coculturing primary CD8 T cells with Tregs only modestly affected the CD8 surfaceome but did partially reverse activation-induced surfaceomic changes. In contrast, hypoxia drastically altered the CD8 surfaceome in a manner consistent with both metabolic reprogramming and induction of an immunosuppressed state. The CD4 T cell surfaceome similarly responded to hypoxia, revealing a common hypoxia-induced surface receptor program. Our surfaceomics findings suggest that hypoxic environments create a challenge for T cell activation. These studies provide global insight into how Tregs and hypoxia remodel the T cell surfaceome and we believe represent a valuable resource to inform future therapeutic efforts to enhance T cell function.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Ashraf, Khuram U; Nygaard, Rie; Vickery, Owen N; Erramilli, Satchal K; Herrera, Carmen M; McConville, Thomas H; Petrou, Vasileios I; Giacometti, Sabrina I; Dufrisne, Meagan Belcher; Nosol, Kamil; Zinkle, Allen P; Graham, Chris L B; Loukeris, Michael; Kloss, Brian; Skorupinska-Tudek, Karolina; Swiezewska, Ewa; Roper, David I; Clarke, Oliver B; Uhlemann, Anne-Catrin; Kossiakoff, Anthony A; Trent, M Stephen; Stansfeld, Phillip J; Mancia, Filippo
Structural basis of lipopolysaccharide maturation by the O-antigen ligase Journal Article
In: Nature, vol. 604, no. 7905, pp. 371–376, 2022, ISSN: 1476-4687.
@article{pmid35388216,
title = {Structural basis of lipopolysaccharide maturation by the O-antigen ligase},
author = {Khuram U Ashraf and Rie Nygaard and Owen N Vickery and Satchal K Erramilli and Carmen M Herrera and Thomas H McConville and Vasileios I Petrou and Sabrina I Giacometti and Meagan Belcher Dufrisne and Kamil Nosol and Allen P Zinkle and Chris L B Graham and Michael Loukeris and Brian Kloss and Karolina Skorupinska-Tudek and Ewa Swiezewska and David I Roper and Oliver B Clarke and Anne-Catrin Uhlemann and Anthony A Kossiakoff and M Stephen Trent and Phillip J Stansfeld and Filippo Mancia},
doi = {10.1038/s41586-022-04555-x},
issn = {1476-4687},
year = {2022},
date = {2022-04-01},
urldate = {2022-04-01},
journal = {Nature},
volume = {604},
number = {7905},
pages = {371--376},
abstract = {The outer membrane of Gram-negative bacteria has an external leaflet that is largely composed of lipopolysaccharide, which provides a selective permeation barrier, particularly against antimicrobials. The final and crucial step in the biosynthesis of lipopolysaccharide is the addition of a species-dependent O-antigen to the lipid A core oligosaccharide, which is catalysed by the O-antigen ligase WaaL. Here we present structures of WaaL from Cupriavidus metallidurans, both in the apo state and in complex with its lipid carrier undecaprenyl pyrophosphate, determined by single-particle cryo-electron microscopy. The structures reveal that WaaL comprises 12 transmembrane helices and a predominantly α-helical periplasmic region, which we show contains many of the conserved residues that are required for catalysis. We observe a conserved fold within the GT-C family of glycosyltransferases and hypothesize that they have a common mechanism for shuttling the undecaprenyl-based carrier to and from the active site. The structures, combined with genetic, biochemical, bioinformatics and molecular dynamics simulation experiments, offer molecular details on how the ligands come in apposition, and allows us to propose a mechanistic model for catalysis. Together, our work provides a structural basis for lipopolysaccharide maturation in a member of the GT-C superfamily of glycosyltransferases.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
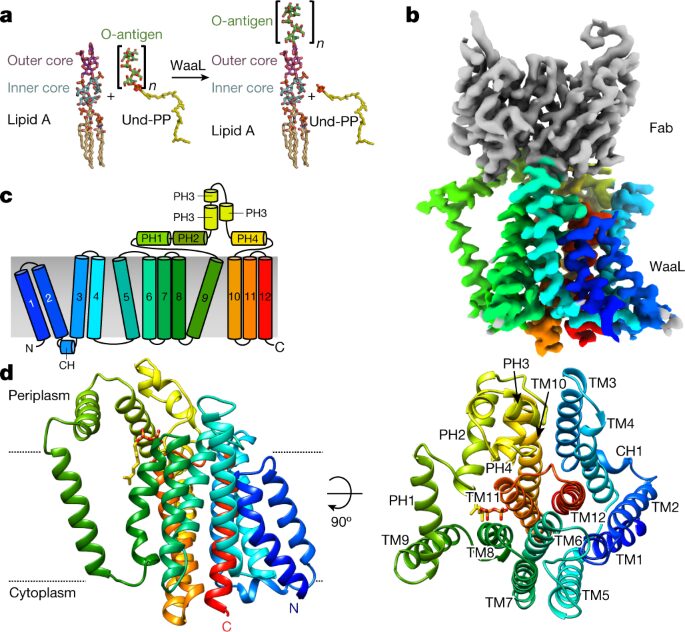
Rohaim, Ahmed; Slezak, Tomasz; Koh, Young Hoon; Blachowicz, Lydia; Kossiakoff, Anthony A; Roux, Benoît
Engineering of a synthetic antibody fragment for structural and functional studies of K+ channels Journal Article
In: J Gen Physiol, vol. 154, no. 4, 2022, ISSN: 1540-7748.
@article{pmid35234830,
title = {Engineering of a synthetic antibody fragment for structural and functional studies of K+ channels},
author = {Ahmed Rohaim and Tomasz Slezak and Young Hoon Koh and Lydia Blachowicz and Anthony A Kossiakoff and Benoît Roux},
doi = {10.1085/jgp.202112965},
issn = {1540-7748},
year = {2022},
date = {2022-04-01},
urldate = {2022-04-01},
journal = {J Gen Physiol},
volume = {154},
number = {4},
abstract = {Engineered antibody fragments (Fabs) have made major impacts on structural biology research, particularly to aid structural determination of membrane proteins. Nonetheless, Fabs generated by traditional monoclonal technology suffer from challenges of routine production and storage. Starting from the known IgG paratopes of an antibody that binds to the "turret loop" of the KcsA K+ channel, we engineered a synthetic Fab (sFab) based upon the highly stable Herceptin Fab scaffold, which can be recombinantly expressed in Escherichia coli and purified with single-step affinity chromatography. This synthetic Fab was used as a crystallization chaperone to obtain crystals of the KcsA channel that diffracted to a resolution comparable to that from the parent Fab. Furthermore, we show that the turret loop can be grafted into the unrelated voltage-gated Kv1.2-Kv2.1 channel and still strongly bind the engineered sFab, in support of the loop grafting strategy. Macroscopic electrophysiology recordings show that the sFab affects the activation and conductance of the chimeric voltage-gated channel. These results suggest that straightforward engineering of antibodies using recombinant formats can facilitate the rapid and scalable production of Fabs as structural biology tools and functional probes. The impact of this approach is expanded significantly based on the potential portability of the turret loop to a myriad of other K+ channels.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
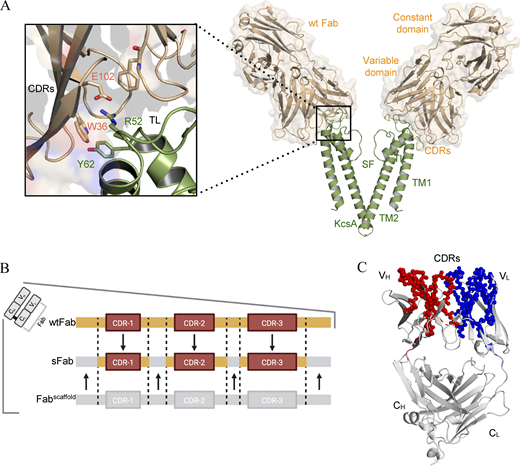
Glasgow, Jeff E.; Byrnes, James R.; Barbee, Susannah D.; Moreau, Joshua M.; Rosenblum, Michael D.; Wells, James A.
Identifying and antagonizing the interactions between layilin and glycosylated collagens Journal Article
In: Cell Chemical Biology, vol. 29, no. 4, pp. 597–604.e7, 2022, ISSN: 2451-9456.
@article{Glasgow2022,
title = {Identifying and antagonizing the interactions between layilin and glycosylated collagens},
author = {Jeff E. Glasgow and James R. Byrnes and Susannah D. Barbee and Joshua M. Moreau and Michael D. Rosenblum and James A. Wells},
doi = {10.1016/j.chembiol.2022.01.003},
issn = {2451-9456},
year = {2022},
date = {2022-04-00},
urldate = {2022-04-00},
journal = {Cell Chemical Biology},
volume = {29},
number = {4},
pages = {597--604.e7},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
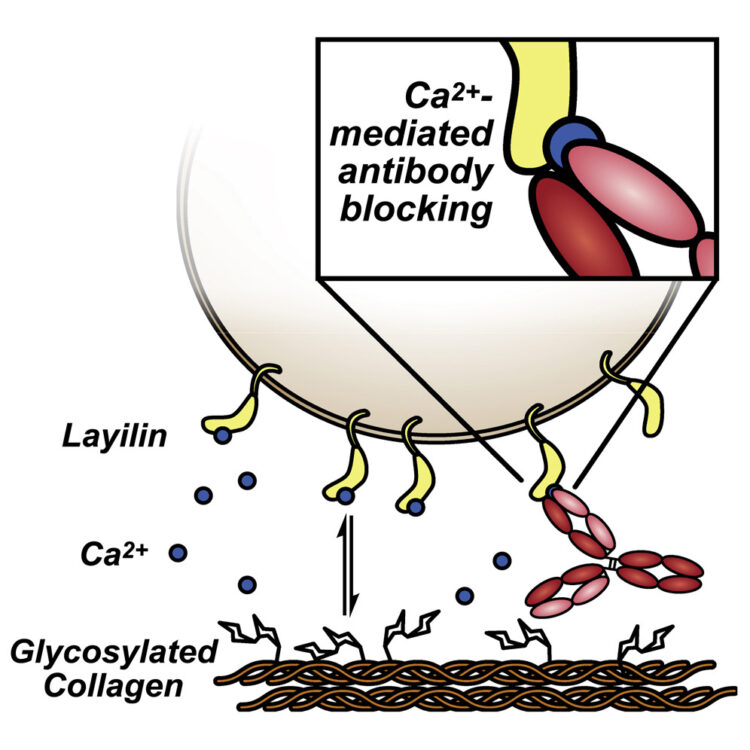
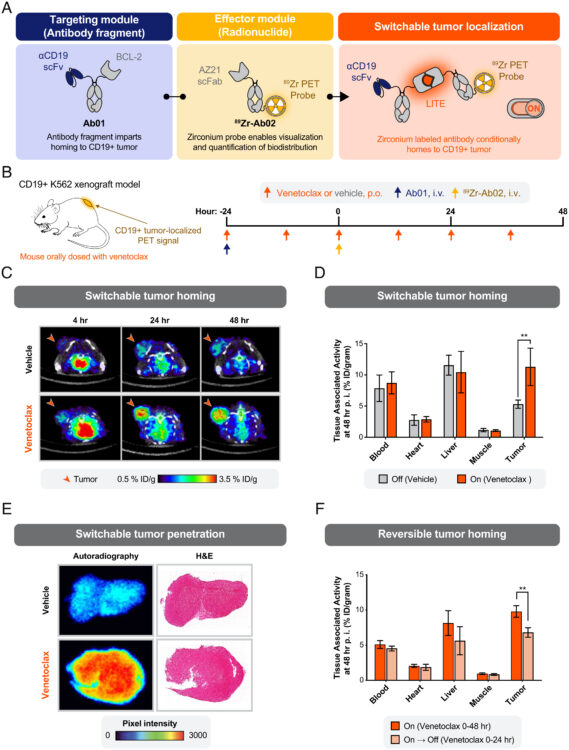
Martinko, Alexander J; Simonds, Erin F; Prasad, Suchitra; Ponce, Alberto; Bracken, Colton J; Wei, Junnian; Wang, Yung-Hua; Chow, Tiffany-Lynn; Huang, Zhong; Evans, Michael J; Wells, James A; Hill, Zachary B
Switchable assembly and function of antibody complexes in vivo using a small molecule Journal Article
In: Proc Natl Acad Sci U S A, vol. 119, no. 9, 2022, ISSN: 1091-6490.
@article{pmid35210365,
title = {Switchable assembly and function of antibody complexes in vivo using a small molecule},
author = {Alexander J Martinko and Erin F Simonds and Suchitra Prasad and Alberto Ponce and Colton J Bracken and Junnian Wei and Yung-Hua Wang and Tiffany-Lynn Chow and Zhong Huang and Michael J Evans and James A Wells and Zachary B Hill},
doi = {10.1073/pnas.2117402119},
issn = {1091-6490},
year = {2022},
date = {2022-03-01},
urldate = {2022-03-01},
journal = {Proc Natl Acad Sci U S A},
volume = {119},
number = {9},
abstract = {The antigen specificity and long serum half-life of monoclonal antibodies have made them a critical part of modern therapeutics. These properties have been coopted in a number of synthetic formats, such as antibody-drug conjugates, bispecific antibodies, or Fc-fusion proteins to generate novel biologic drug modalities. Historically, these new therapies have been generated by covalently linking multiple molecular moieties through chemical or genetic methods. This irreversible fusion of different components means that the function of the molecule is static, as determined by the structure. Here, we report the development of a technology for switchable assembly of functional antibody complexes using chemically induced dimerization domains. This approach enables control of the antibody's intended function in vivo by modulating the dose of a small molecule. We demonstrate this switchable assembly across three therapeutically relevant functionalities in vivo, including localization of a radionuclide-conjugated antibody to an antigen-positive tumor, extension of a cytokine's half-life, and activation of bispecific, T cell-engaging antibodies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Kirkemo, Lisa L; Elledge, Susanna K; Yang, Jiuling; Byrnes, James R; Glasgow, Jeff E; Blelloch, Robert; Wells, James A
In: Elife, vol. 11, 2022, ISSN: 2050-084X.
@article{pmid35257663,
title = {Cell-surface tethered promiscuous biotinylators enable comparative small-scale surface proteomic analysis of human extracellular vesicles and cells},
author = {Lisa L Kirkemo and Susanna K Elledge and Jiuling Yang and James R Byrnes and Jeff E Glasgow and Robert Blelloch and James A Wells},
doi = {10.7554/eLife.73982},
issn = {2050-084X},
year = {2022},
date = {2022-03-01},
urldate = {2022-03-01},
journal = {Elife},
volume = {11},
abstract = {Characterization of cell surface proteome differences between cancer and healthy cells is a valuable approach for the identification of novel diagnostic and therapeutic targets. However, selective sampling of surface proteins for proteomics requires large samples (>10e6 cells) and long labeling times. These limitations preclude analysis of material-limited biological samples or the capture of rapid surface proteomic changes. Here, we present two labeling approaches to tether exogenous peroxidases (APEX2 and HRP) directly to cells, enabling rapid, small-scale cell surface biotinylation without the need to engineer cells. We used a novel lipidated DNA-tethered APEX2 (DNA-APEX2), which upon addition to cells promoted cell agnostic membrane-proximal labeling. Alternatively, we employed horseradish peroxidase (HRP) fused to the glycan-binding domain of wheat germ agglutinin (WGA-HRP). This approach yielded a rapid and commercially inexpensive means to directly label cells containing common N-Acetylglucosamine (GlcNAc) and sialic acid glycans on their surface. The facile WGA-HRP method permitted high surface coverage of cellular samples and enabled the first comparative surface proteome characterization of cells and cell-derived small extracellular vesicles (EVs), leading to the robust quantification of 953 cell and EV surface annotated proteins. We identified a newly recognized subset of EV-enriched markers, as well as proteins that are uniquely upregulated on Myc oncogene-transformed prostate cancer EVs. These two cell-tethered enzyme surface biotinylation approaches are highly advantageous for rapidly and directly labeling surface proteins across a range of material-limited sample types.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
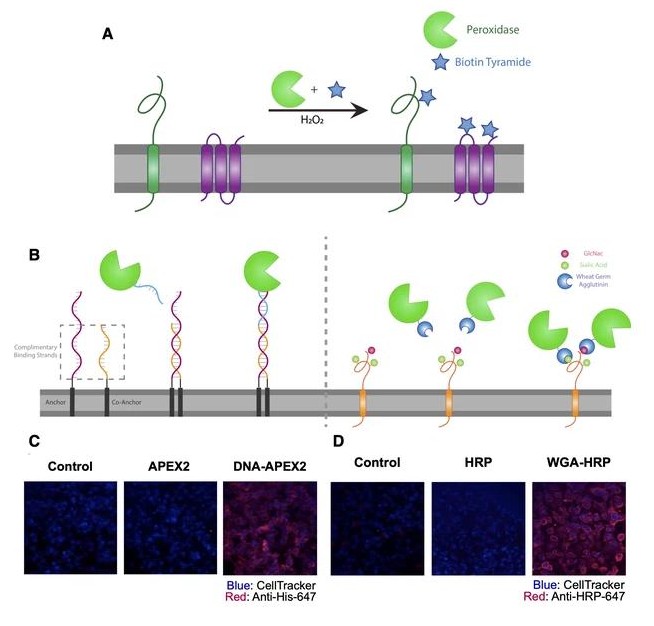
Rettko, Nicholas J; Campisi, Judith; Wells, James A
Engineering Antibodies Targeting p16 MHC-Peptide Complexes Journal Article
In: ACS Chem Biol, vol. 17, no. 3, pp. 545–555, 2022, ISSN: 1554-8937.
@article{pmid35212540,
title = {Engineering Antibodies Targeting p16 MHC-Peptide Complexes},
author = {Nicholas J Rettko and Judith Campisi and James A Wells},
doi = {10.1021/acschembio.1c00808},
issn = {1554-8937},
year = {2022},
date = {2022-03-01},
urldate = {2022-03-01},
journal = {ACS Chem Biol},
volume = {17},
number = {3},
pages = {545--555},
abstract = {Senescent cells undergo a permanent cell cycle arrest and drive a host of age-related pathologies. Recent transgenic mouse models indicate that removing cells expressing the senescence marker p16 (p16) can increase median lifespan and delay the onset of many aging phenotypes. However, identifying and eliminating native human cells expressing p16 has remained a challenge. We hypothesize that senescent cells display peptides derived from p16 in major histocompatibility complex (MHC)-peptide complexes on the cell surface that could serve as targetable antigens for antibody-based biologics. Using Fab-phage display technology, we generated antibodies that bind to a p16 MHC-peptide complex from the human leukocyte antigen (HLA) allele HLA-B*35:01. When converted to single-chain Fab chimeric antigen receptor (CAR) constructs, these antibodies can recognize naturally presented p16 MHC-peptide complexes on the surface of cells and activate Jurkat cells. Furthermore, we developed antibodies against predicted p16 MHC-peptide complexes for HLA-A*02:01 that specifically recognize their respective antigen on the surface of cells. These tools establish a platform to survey the surface of senescent cells and provide a potential novel senolytic strategy.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
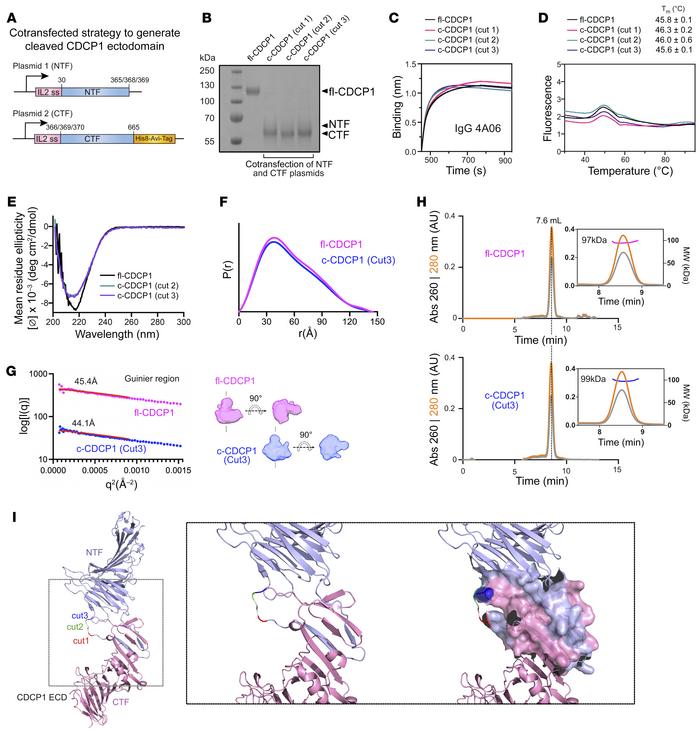
Lim, Shion A; Zhou, Jie; Martinko, Alexander J; Wang, Yung-Hua; Filippova, Ekaterina V; Steri, Veronica; Wang, Donghui; Remesh, Soumya G; Liu, Jia; Hann, Byron; Kossiakoff, Anthony A; Evans, Michael J; Leung, Kevin K; Wells, James A
Targeting a proteolytic neoepitope on CUB domain containing protein 1 (CDCP1) for RAS-driven cancers Journal Article
In: J Clin Invest, vol. 132, no. 4, 2022, ISSN: 1558-8238.
@article{pmid35166238,
title = {Targeting a proteolytic neoepitope on CUB domain containing protein 1 (CDCP1) for RAS-driven cancers},
author = {Shion A Lim and Jie Zhou and Alexander J Martinko and Yung-Hua Wang and Ekaterina V Filippova and Veronica Steri and Donghui Wang and Soumya G Remesh and Jia Liu and Byron Hann and Anthony A Kossiakoff and Michael J Evans and Kevin K Leung and James A Wells},
doi = {10.1172/JCI154604},
issn = {1558-8238},
year = {2022},
date = {2022-02-01},
urldate = {2022-02-01},
journal = {J Clin Invest},
volume = {132},
number = {4},
abstract = {Extracellular proteolysis is frequently dysregulated in disease and can generate proteoforms with unique neoepitopes not found in healthy tissue. Here, we demonstrate that Abs that selectively recognize a proteolytic neoepitope on CUB domain containing protein 1 (CDCP1) could enable more effective and safer treatments for solid tumors. CDCP1 is highly overexpressed in RAS-driven cancers, and its ectodomain is cleaved by extracellular proteases. Biochemical, biophysical, and structural characterization revealed that the 2 cleaved fragments of CDCP1 remain tightly associated with minimal proteolysis-induced conformational change. Using differential phage display, we generated recombinant Abs that are exquisitely selective to cleaved CDCP1 with no detectable binding to the uncleaved form. These Abs potently targeted cleaved CDCP1-expressing cancer cells as an Ab-drug conjugate, an Ab-radionuclide conjugate, and a bispecific T cell engager. In a syngeneic pancreatic tumor model, these cleaved-specific Abs showed tumor-specific localization and antitumor activity with superior safety profiles compared with a pan-CDCP1 approach. Targeting proteolytic neoepitopes could provide an orthogonal "AND" gate for improving the therapeutic index.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Safaee, Michael M; Wang, Elaina J; Jain, Saket; Chen, Jia-Shu; Gill, Sabraj; Zheng, Allison C; Garcia, Joseph H; Beniwal, Angad S; Tran, Y; Nguyen, Alan T; Trieu, Melissa; Leung, Kevin; Wells, Jim; Maclean, James M; Wycoff, Keith; Aghi, Manish K
CD97 is associated with mitogenic pathway activation, metabolic reprogramming, and immune microenvironment changes in glioblastoma Journal Article
In: Sci Rep, vol. 12, no. 1, pp. 1464, 2022, ISSN: 2045-2322.
@article{pmid35087132,
title = {CD97 is associated with mitogenic pathway activation, metabolic reprogramming, and immune microenvironment changes in glioblastoma},
author = {Michael M Safaee and Elaina J Wang and Saket Jain and Jia-Shu Chen and Sabraj Gill and Allison C Zheng and Joseph H Garcia and Angad S Beniwal and Y Tran and Alan T Nguyen and Melissa Trieu and Kevin Leung and Jim Wells and James M Maclean and Keith Wycoff and Manish K Aghi},
doi = {10.1038/s41598-022-05259-y},
issn = {2045-2322},
year = {2022},
date = {2022-01-01},
urldate = {2022-01-01},
journal = {Sci Rep},
volume = {12},
number = {1},
pages = {1464},
abstract = {Glioblastoma (GBM) is the most common primary brain tumor with a median survival under two years. Using in silico and in vitro techniques, we demonstrate heterogeneous expression of CD97, a leukocyte adhesion marker, in human GBM. Beyond its previous demonstrated role in tumor invasion, we show that CD97 is also associated with upregulation of the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/Erk) and phosphatidylinositol 3-kinase/protein kinase B (PI3K/Akt) pathways in GBM. While CD97 knockout decreased Akt activation, CD97 targeting did not alter MAPK/Erk activation, did not slow GBM cell proliferation in culture, and increased levels of glycolytic and oxidative phosphorylation metabolites. Treatment with a soluble CD97 inhibitor did not alter activation of the MAPK/Erk and PI3K/Akt pathways. Tumors with high CD97 expression were associated with immune microenvironment changes including increased naïve macrophages, regulatory T cells, and resting natural killer (NK) cells. These data suggest that, while CD97 expression is associated with conflicting effects on tumor cell proliferative and metabolic pathways that overall do not affect tumor cell proliferation, CD97 exerts pro-tumoral effects on the tumor immune microenvironment, which along with the pro-invasive effects of CD97 we previously demonstrated, provides impetus to continue exploring CD97 as a therapeutic target in GBM.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}