Publications: 2021
2021
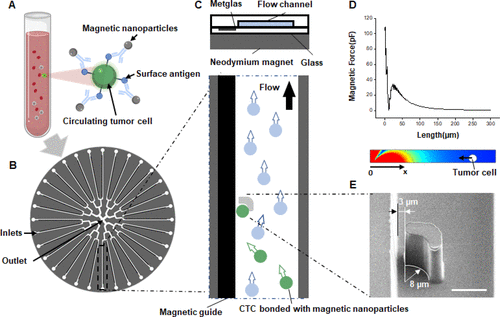
Ma, Yuan; Chen, Kangfu; Xia, Fan; Atwal, Randy; Wang, Hansen; Ahmed, Sharif U; Cardarelli, Lia; Lui, Irene; Duong, Bill; Wang, Zongjie; Wells, James A; Sidhu, Sachdev S; Kelley, Shana O
Phage-Based Profiling of Rare Single Cells Using Nanoparticle-Directed Capture Journal Article
In: ACS Nano, vol. 15, no. 12, pp. 19202–19210, 2021, ISSN: 1936-086X.
@article{pmid34813293,
title = {Phage-Based Profiling of Rare Single Cells Using Nanoparticle-Directed Capture},
author = {Yuan Ma and Kangfu Chen and Fan Xia and Randy Atwal and Hansen Wang and Sharif U Ahmed and Lia Cardarelli and Irene Lui and Bill Duong and Zongjie Wang and James A Wells and Sachdev S Sidhu and Shana O Kelley},
doi = {10.1021/acsnano.1c03935},
issn = {1936-086X},
year = {2021},
date = {2021-12-01},
urldate = {2021-12-01},
journal = {ACS Nano},
volume = {15},
number = {12},
pages = {19202--19210},
abstract = {Advances in single-cell level profiling of the proteome require quantitative and versatile platforms, especially for rare cell analyses such as circulating tumor cell (CTC) profiling. Here we demonstrate an integrated microfluidic chip that uses magnetic nanoparticles to capture single tumor cells with high efficiency, permits on-chip incubation, and facilitates cell-surface protein expression analysis. Combined with phage-based barcoding and next-generation sequencing technology, we were able to monitor changes in the expression of multiple surface markers stimulated in response to CTC adherence. Interestingly, we found fluctuations in the expression of Frizzled2 (FZD2) that reflected the microenvironment of the single cells. This platform has a high potential for in-depth screening of multiple surface antigens simultaneously in rare cells with single-cell resolution, which will provide further insights regarding biological heterogeneity and human disease.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
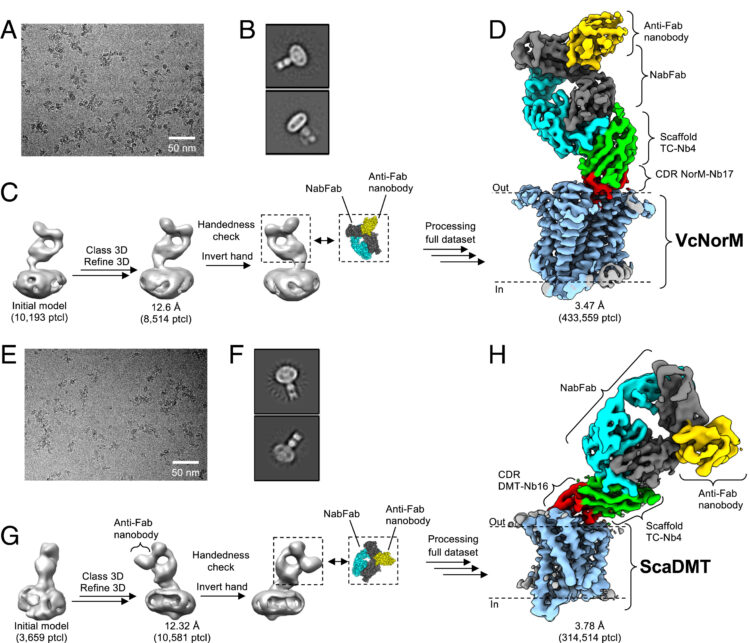
Bloch, Joël S; Mukherjee, Somnath; Kowal, Julia; Filippova, Ekaterina V; Niederer, Martina; Pardon, Els; Steyaert, Jan; Kossiakoff, Anthony A; Locher, Kaspar P
Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins Journal Article
In: Proc Natl Acad Sci U S A, vol. 118, no. 47, 2021, ISSN: 1091-6490.
@article{pmid34782475,
title = {Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins},
author = {Joël S Bloch and Somnath Mukherjee and Julia Kowal and Ekaterina V Filippova and Martina Niederer and Els Pardon and Jan Steyaert and Anthony A Kossiakoff and Kaspar P Locher},
doi = {10.1073/pnas.2115435118},
issn = {1091-6490},
year = {2021},
date = {2021-11-01},
urldate = {2021-11-01},
journal = {Proc Natl Acad Sci U S A},
volume = {118},
number = {47},
abstract = {With conformation-specific nanobodies being used for a wide range of structural, biochemical, and cell biological applications, there is a demand for antigen-binding fragments (Fabs) that specifically and tightly bind these nanobodies without disturbing the nanobody-target protein interaction. Here, we describe the development of a synthetic Fab (termed NabFab) that binds the scaffold of an alpaca-derived nanobody with picomolar affinity. We demonstrate that upon complementary-determining region grafting onto this parent nanobody scaffold, nanobodies recognizing diverse target proteins and derived from llama or camel can cross-react with NabFab without loss of affinity. Using NabFab as a fiducial and size enhancer (50 kDa), we determined the high-resolution cryogenic electron microscopy (cryo-EM) structures of nanobody-bound VcNorM and ScaDMT, both small membrane proteins of ∼50 kDa. Using an additional anti-Fab nanobody further facilitated reliable initial three-dimensional structure determination from small cryo-EM test datasets. Given that NabFab is of synthetic origin, is humanized, and can be conveniently expressed in in large amounts, it may be useful not only for structural biology but also for biomedical applications.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
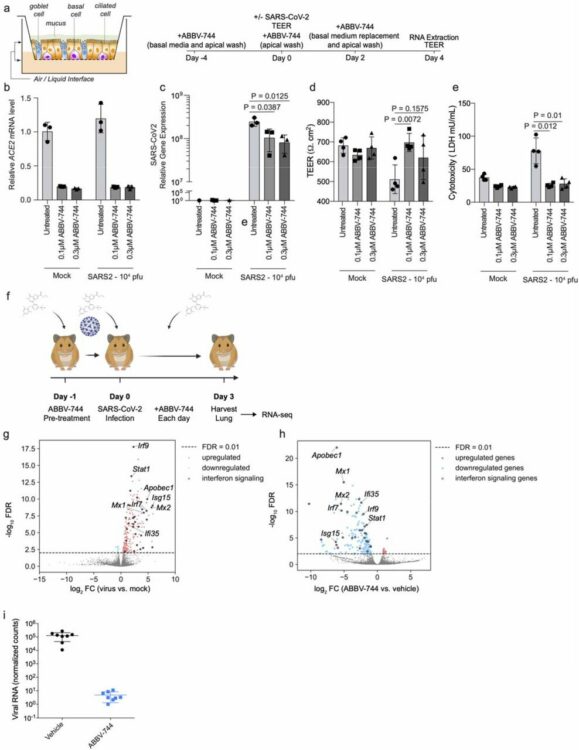
Samelson, Avi J; Tran, Quang Dinh; Robinot, Rémy; Carrau, Lucia; Rezelj, Veronica V; Kain, Alice Mac; Chen, Merissa; Ramadoss, Gokul N; Guo, Xiaoyan; Lim, Shion A; Lui, Irene; Nunez, James; Rockwood, Sarah J; Wang, Jianhui; Liu, Na; Carlson-Stevermer, Jared; Oki, Jennifer; Maures, Travis; Holden, Kevin; Weissman, Jonathan S; Wells, James A; Conklin, Bruce R; TenOever, Benjamin R; Chakrabarti, Lisa A; Vignuzzi, Marco; Tian, Ruilin; Kampmann, Martin
BRD2 inhibition blocks SARS-CoV-2 infection by reducing transcription of the host cell receptor ACE2 Journal Article
In: bioRxiv, 2021, ISSN: 2692-8205.
@article{pmid33501440,
title = {BRD2 inhibition blocks SARS-CoV-2 infection by reducing transcription of the host cell receptor ACE2},
author = {Avi J Samelson and Quang Dinh Tran and Rémy Robinot and Lucia Carrau and Veronica V Rezelj and Alice Mac Kain and Merissa Chen and Gokul N Ramadoss and Xiaoyan Guo and Shion A Lim and Irene Lui and James Nunez and Sarah J Rockwood and Jianhui Wang and Na Liu and Jared Carlson-Stevermer and Jennifer Oki and Travis Maures and Kevin Holden and Jonathan S Weissman and James A Wells and Bruce R Conklin and Benjamin R TenOever and Lisa A Chakrabarti and Marco Vignuzzi and Ruilin Tian and Martin Kampmann},
doi = {10.1101/2021.01.19.427194},
issn = {2692-8205},
year = {2021},
date = {2021-09-01},
urldate = {2021-09-01},
journal = {bioRxiv},
abstract = {SARS-CoV-2 infection of human cells is initiated by the binding of the viral Spike protein to its cell-surface receptor ACE2. We conducted a targeted CRISPRi screen to uncover druggable pathways controlling Spike protein binding to human cells. We found that the protein BRD2 is required for transcription in human lung epithelial cells and cardiomyocytes, and BRD2 inhibitors currently evaluated in clinical trials potently block endogenous expression and SARS-CoV-2 infection of human cells, including those of human nasal epithelia. Moreover, pharmacological BRD2 inhibition with the drug ABBV-744 inhibited SARS-CoV-2 replication in Syrian hamsters. We also found that BRD2 controls transcription of several other genes induced upon SARS-CoV-2 infection, including the interferon response, which in turn regulates the antiviral response. Together, our results pinpoint BRD2 as a potent and essential regulator of the host response to SARS-CoV-2 infection and highlight the potential of BRD2 as a novel therapeutic target for COVID-19.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
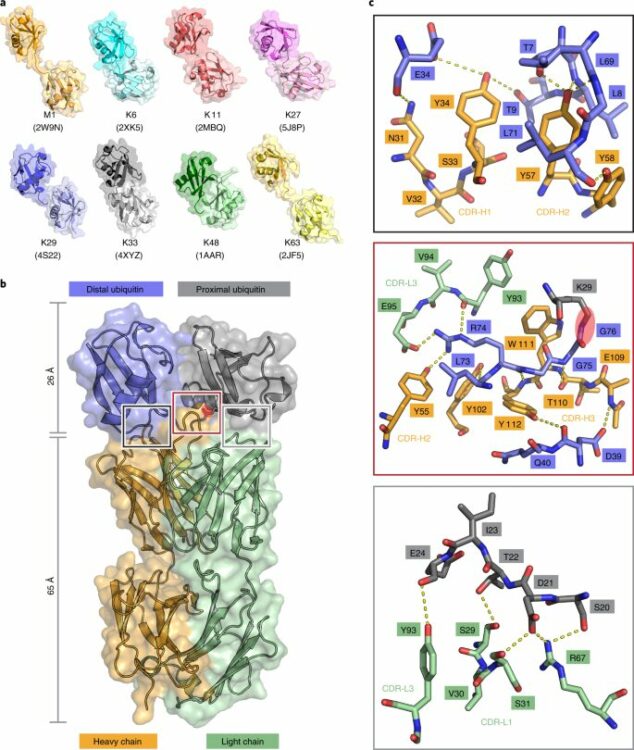
Yu, Yuanyuan; Zheng, Qingyun; Erramilli, Satchal K; Pan, Man; Park, Seongjin; Xie, Yuan; Li, Jingxian; Fei, Jingyi; Kossiakoff, Anthony A; Liu, Lei; Zhao, Minglei
K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle Journal Article
In: Nat Chem Biol, vol. 17, no. 8, pp. 896–905, 2021, ISSN: 1552-4469.
@article{pmid34239127,
title = {K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle},
author = {Yuanyuan Yu and Qingyun Zheng and Satchal K Erramilli and Man Pan and Seongjin Park and Yuan Xie and Jingxian Li and Jingyi Fei and Anthony A Kossiakoff and Lei Liu and Minglei Zhao},
doi = {10.1038/s41589-021-00823-5},
issn = {1552-4469},
year = {2021},
date = {2021-08-01},
urldate = {2021-08-01},
journal = {Nat Chem Biol},
volume = {17},
number = {8},
pages = {896--905},
abstract = {Protein ubiquitination shows remarkable topological and functional diversity through the polymerization of ubiquitin via different linkages. Deciphering the cellular ubiquitin code is of central importance to understand the physiology of the cell. However, our understanding of its function is rather limited due to the lack of specific binders as tools to detect K29-linked polyubiquitin. In this study, we screened and characterized a synthetic antigen-binding fragment, termed sAB-K29, that can specifically recognize K29-linked polyubiquitin using chemically synthesized K29-linked diubiquitin. We further determined the crystal structure of this fragment bound to the K29-linked diubiquitin, which revealed the molecular basis of specificity. Using sAB-K29 as a tool, we uncovered that K29-linked ubiquitination is involved in different kinds of cellular proteotoxic stress response as well as cell cycle regulation. In particular, we showed that K29-linked ubiquitination is enriched in the midbody and downregulation of the K29-linked ubiquitination signal arrests cells in G1/S phase.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
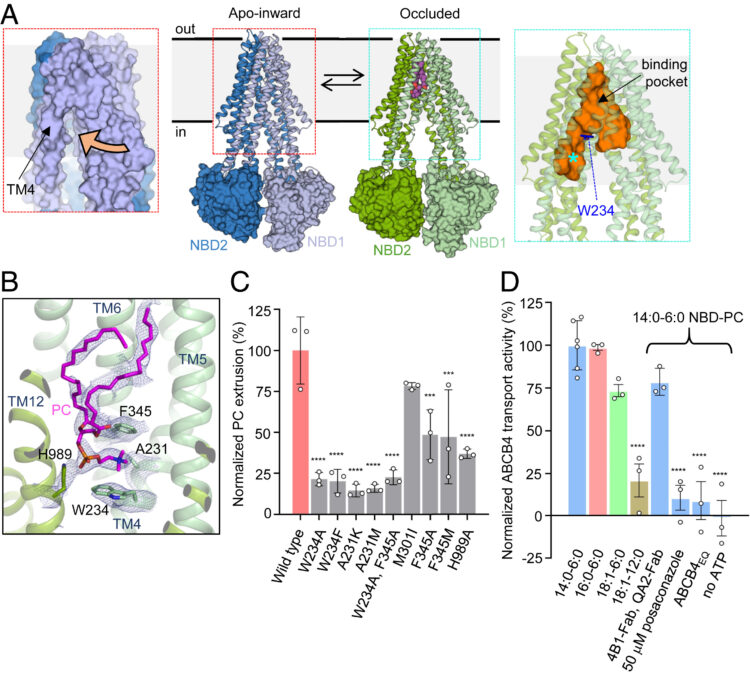
Nosol, Kamil; Bang-Sørensen, Rose; Irobalieva, Rossitza N; Erramilli, Satchal K; Stieger, Bruno; Kossiakoff, Anthony A; Locher, Kaspar P
Structures of ABCB4 provide insight into phosphatidylcholine translocation Journal Article
In: Proc Natl Acad Sci U S A, vol. 118, no. 33, 2021, ISSN: 1091-6490.
@article{pmid34385322,
title = {Structures of ABCB4 provide insight into phosphatidylcholine translocation},
author = {Kamil Nosol and Rose Bang-Sørensen and Rossitza N Irobalieva and Satchal K Erramilli and Bruno Stieger and Anthony A Kossiakoff and Kaspar P Locher},
doi = {10.1073/pnas.2106702118},
issn = {1091-6490},
year = {2021},
date = {2021-08-01},
urldate = {2021-08-01},
journal = {Proc Natl Acad Sci U S A},
volume = {118},
number = {33},
abstract = {ABCB4 is expressed in hepatocytes and translocates phosphatidylcholine into bile canaliculi. The mechanism of specific lipid recruitment from the canalicular membrane, which is essential to mitigate the cytotoxicity of bile salts, is poorly understood. We present cryogenic electron microscopy structures of human ABCB4 in three distinct functional conformations. An apo-inward structure reveals how phospholipid can be recruited from the inner leaflet of the membrane without flipping its orientation. An occluded structure reveals a single phospholipid molecule in a central cavity. Its choline moiety is stabilized by cation-π interactions with an essential tryptophan residue, rationalizing the specificity of ABCB4 for phosphatidylcholine. In an inhibitor-bound structure, a posaconazole molecule blocks phospholipids from reaching the central cavity. Using a proteoliposome-based translocation assay with fluorescently labeled phosphatidylcholine analogs, we recapitulated the substrate specificity of ABCB4 in vitro and confirmed the role of the key tryptophan residue. Our results provide a structural basis for understanding an essential translocation step in the generation of bile and its sensitivity to azole drugs.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
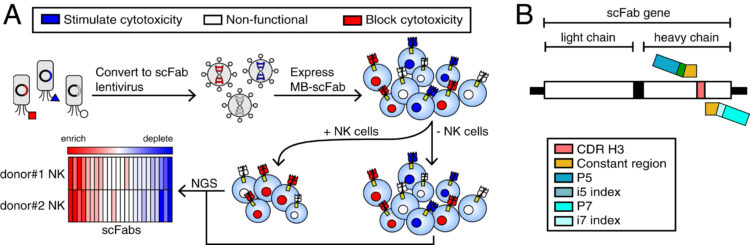
Kang, Emily; Kadoch, Cigall; Rubenstein, James L; Lanier, Lewis L; Wells, James A
A functional mammalian display screen identifies rare antibodies that stimulate NK cell-mediated cytotoxicity Journal Article
In: Proc Natl Acad Sci U S A, vol. 118, no. 31, 2021, ISSN: 1091-6490.
@article{pmid34330834,
title = {A functional mammalian display screen identifies rare antibodies that stimulate NK cell-mediated cytotoxicity},
author = {Emily Kang and Cigall Kadoch and James L Rubenstein and Lewis L Lanier and James A Wells},
doi = {10.1073/pnas.2104099118},
issn = {1091-6490},
year = {2021},
date = {2021-08-01},
urldate = {2021-08-01},
journal = {Proc Natl Acad Sci U S A},
volume = {118},
number = {31},
abstract = {Therapies that boost the antitumor immune response have shown a great deal of success. Although most of these therapies have focused on enhancing T cell functions, there is a growing interest in developing therapies that can target other immune cell subsets. Like T cells, natural killer (NK) cells are cytotoxic effector cells that play a key role in the antitumor response. To advance the development of NK-based therapies, we developed a functional screen to rapidly identify antibodies that can activate NK cells. We displayed antibodies on a mammalian target cell line and probed their ability to stimulate NK cell-mediated cytotoxicity. From this screen, we identified five antibodies that bound with high affinity to NK cells and stimulated NK cell-mediated cytotoxicity and interferon-γ (IFN-γ) secretion. We demonstrate that these antibodies can be further developed into bispecific antibodies to redirect NK cell-mediated cytotoxicity toward CD20+ B cell lymphoma cells and HER2+ breast cancer cells. While antibodies to two of the receptors, CD16 and NCR1, have previously been targeted as bispecific antibodies to redirect NK cell-mediated cytotoxicity, we demonstrate that bispecific antibodies targeting NCR3 can also potently activate NK cells. These results show that this screen can be used to directly identify antibodies that can enhance antitumor immune responses.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
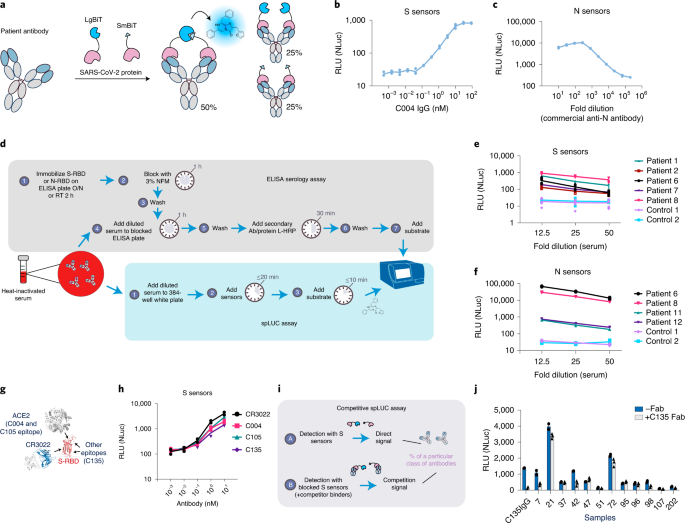
Elledge, Susanna K.; Zhou, Xin X.; Byrnes, James R.; Martinko, Alexander J.; Lui, Irene; Pance, Katarina; Lim, Shion A.; Glasgow, Jeff E.; Glasgow, Anum A.; Turcios, Keirstinne; Iyer, Nikita S.; Torres, Leonel; Peluso, Michael J.; Henrich, Timothy J.; Wang, Taia T.; Tato, Cristina M.; Leung, Kevin K.; Greenhouse, Bryan; Wells, James A.
Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection Journal Article
In: Nat Biotechnol, vol. 39, no. 8, pp. 928–935, 2021, ISSN: 1546-1696.
@article{Elledge2021,
title = {Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection},
author = {Susanna K. Elledge and Xin X. Zhou and James R. Byrnes and Alexander J. Martinko and Irene Lui and Katarina Pance and Shion A. Lim and Jeff E. Glasgow and Anum A. Glasgow and Keirstinne Turcios and Nikita S. Iyer and Leonel Torres and Michael J. Peluso and Timothy J. Henrich and Taia T. Wang and Cristina M. Tato and Kevin K. Leung and Bryan Greenhouse and James A. Wells},
doi = {10.1038/s41587-021-00878-8},
issn = {1546-1696},
year = {2021},
date = {2021-08-00},
urldate = {2021-08-00},
journal = {Nat Biotechnol},
volume = {39},
number = {8},
pages = {928--935},
publisher = {Springer Science and Business Media LLC},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Gallo, Eugenio; Kelil, Abdellali; Haughey, Michael; Cazares-Olivera, Mariana; Yates, Bradley P; Zhang, Mingjun; Wang, Nai-Yu; Blazer, Levi; Carderelli, Lia; Adams, Jarrett J; Kossiakoff, Anthony A; Wells, James A; Xie, Weilin; Sidhu, Sachdev S
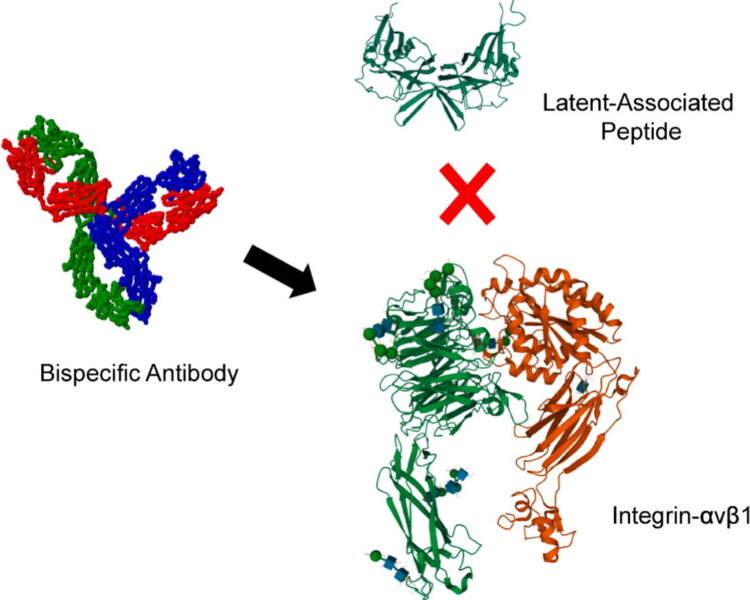
Inhibition of Cancer Cell Adhesion, Migration and Proliferation by a Bispecific Antibody that Targets two Distinct Epitopes on αv Integrins Journal Article
In: J Mol Biol, vol. 433, no. 15, pp. 167090, 2021, ISSN: 1089-8638.
@article{pmid34090922,
title = {Inhibition of Cancer Cell Adhesion, Migration and Proliferation by a Bispecific Antibody that Targets two Distinct Epitopes on αv Integrins},
author = {Eugenio Gallo and Abdellali Kelil and Michael Haughey and Mariana Cazares-Olivera and Bradley P Yates and Mingjun Zhang and Nai-Yu Wang and Levi Blazer and Lia Carderelli and Jarrett J Adams and Anthony A Kossiakoff and James A Wells and Weilin Xie and Sachdev S Sidhu},
doi = {10.1016/j.jmb.2021.167090},
issn = {1089-8638},
year = {2021},
date = {2021-07-01},
urldate = {2021-07-01},
journal = {J Mol Biol},
volume = {433},
number = {15},
pages = {167090},
abstract = {Members of the αv family of integrins regulate activation of transforming growth factor beta (TGFβ) and are directly involved in pro-tumorigenic phenotypes. Thus, αv integrins may be therapeutic targets for fibrosis and cancer, yet the isolation of selective inhibitors is currently a challenge. We generated synthetic antibodies selective for αv integrins by phage display selections on cell lines that displayed integrin heterodimers. We identified antibodies that targeted two distinct epitopes on cell-surface αv integrins and partially inhibited cell adhesion mediated by interactions between integrins and the latency-associated peptide, part of the pro-form of TGFβ. Using the isolated antibody paratope sequences we engineered a bispecific antibody capable of binding to both epitopes simultaneously; this antibody potently and completely inhibited cell adhesion mediated by integrins αvβ1, αvβ3 and αvβ5. In addition, the bispecific antibody inhibited proliferation and migration of lung carcinoma lines, where the highest and lowest potencies observed correlated with integrin-αv cell surface expression levels. Taken together, our results demonstrate that phage display selections with live cells can yield high quality anti-integrin antibodies, which we used as biparatopic building blocks to construct a bispecific antibody that strongly inhibited integrin function and may be a therapeutic candidate for cancer and fibrosis.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
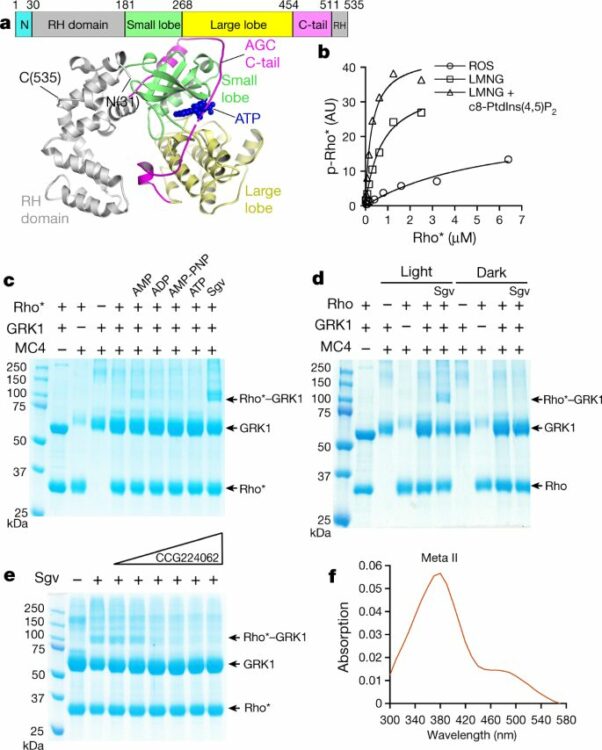
Chen, Qiuyan; Plasencia, Manolo; Li, Zhuang; Mukherjee, Somnath; Patra, Dhabaleswar; Chen, Chun-Liang; Klose, Thomas; Yao, Xin-Qiu; Kossiakoff, Anthony A; Chang, Leifu; Andrews, Philip C; Tesmer, John J G
Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1 Journal Article
In: Nature, vol. 595, no. 7868, pp. 600–605, 2021, ISSN: 1476-4687.
@article{pmid34262173,
title = {Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1},
author = {Qiuyan Chen and Manolo Plasencia and Zhuang Li and Somnath Mukherjee and Dhabaleswar Patra and Chun-Liang Chen and Thomas Klose and Xin-Qiu Yao and Anthony A Kossiakoff and Leifu Chang and Philip C Andrews and John J G Tesmer},
doi = {10.1038/s41586-021-03721-x},
issn = {1476-4687},
year = {2021},
date = {2021-07-01},
urldate = {2021-07-01},
journal = {Nature},
volume = {595},
number = {7868},
pages = {600--605},
abstract = {G-protein-coupled receptor (GPCR) kinases (GRKs) selectively phosphorylate activated GPCRs, thereby priming them for desensitization. Although it is unclear how GRKs recognize these receptors, a conserved region at the GRK N terminus is essential for this process. Here we report a series of cryo-electron microscopy single-particle reconstructions of light-activated rhodopsin (Rho*) bound to rhodopsin kinase (GRK1), wherein the N terminus of GRK1 forms a helix that docks into the open cytoplasmic cleft of Rho*. The helix also packs against the GRK1 kinase domain and stabilizes it in an active configuration. The complex is further stabilized by electrostatic interactions between basic residues that are conserved in most GPCRs and acidic residues that are conserved in GRKs. We did not observe any density for the regulator of G-protein signalling homology domain of GRK1 or the C terminus of rhodopsin. Crosslinking with mass spectrometry analysis confirmed these results and revealed dynamic behaviour in receptor-bound GRK1 that would allow the phosphorylation of multiple sites in the receptor tail. We have identified GRK1 residues whose mutation augments kinase activity and crosslinking with Rho*, as well as residues that are involved in activation by acidic phospholipids. From these data, we present a general model for how a small family of protein kinases can recognize and be activated by hundreds of different GPCRs.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
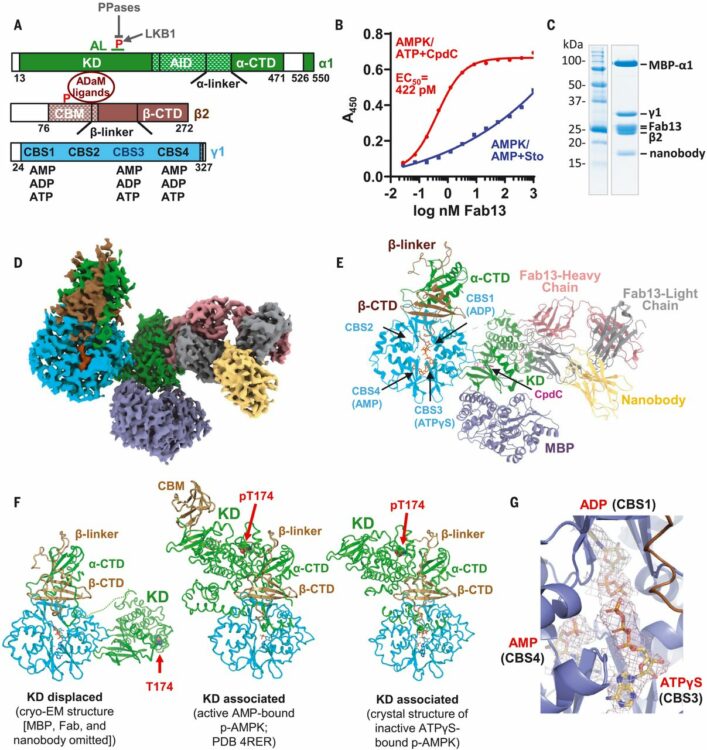
Yan, Yan; Mukherjee, Somnath; Harikumar, Kaleeckal G; Strutzenberg, Timothy S; Zhou, X Edward; Suino-Powell, Kelly; Xu, Ting-Hai; Sheldon, Ryan D; Lamp, Jared; Brunzelle, Joseph S; Radziwon, Katarzyna; Ellis, Abigail; Novick, Scott J; Vega, Irving E; Jones, Russell G; Miller, Laurence J; Xu, H Eric; Griffin, Patrick R; Kossiakoff, Anthony A; Melcher, Karsten
Structure of an AMPK complex in an inactive, ATP-bound state Journal Article
In: Science, vol. 373, no. 6553, pp. 413–419, 2021, ISSN: 1095-9203.
@article{pmid34437114,
title = {Structure of an AMPK complex in an inactive, ATP-bound state},
author = {Yan Yan and Somnath Mukherjee and Kaleeckal G Harikumar and Timothy S Strutzenberg and X Edward Zhou and Kelly Suino-Powell and Ting-Hai Xu and Ryan D Sheldon and Jared Lamp and Joseph S Brunzelle and Katarzyna Radziwon and Abigail Ellis and Scott J Novick and Irving E Vega and Russell G Jones and Laurence J Miller and H Eric Xu and Patrick R Griffin and Anthony A Kossiakoff and Karsten Melcher},
doi = {10.1126/science.abe7565},
issn = {1095-9203},
year = {2021},
date = {2021-07-01},
urldate = {2021-07-01},
journal = {Science},
volume = {373},
number = {6553},
pages = {413--419},
abstract = {Adenosine monophosphate (AMP)-activated protein kinase (AMPK) regulates metabolism in response to the cellular energy states. Under energy stress, AMP stabilizes the active AMPK conformation, in which the kinase activation loop (AL) is protected from protein phosphatases, thus keeping the AL in its active, phosphorylated state. At low AMP:ATP (adenosine triphosphate) ratios, ATP inhibits AMPK by increasing AL dynamics and accessibility. We developed conformation-specific antibodies to trap ATP-bound AMPK in a fully inactive, dynamic state and determined its structure at 3.5-angstrom resolution using cryo-electron microscopy. A 180° rotation and 100-angstrom displacement of the kinase domain fully exposes the AL. On the basis of the structure and supporting biophysical data, we propose a multistep mechanism explaining how adenine nucleotides and pharmacological agonists modulate AMPK activity by altering AL phosphorylation and accessibility.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
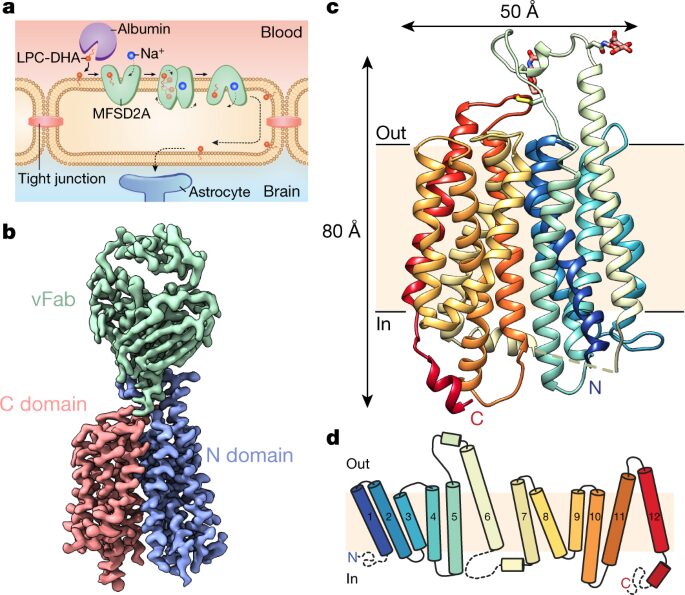
Cater, Rosemary J; Chua, Geok Lin; Erramilli, Satchal K; Keener, James E; Choy, Brendon C; Tokarz, Piotr; Chin, Cheen Fei; Quek, Debra Q Y; Kloss, Brian; Pepe, Joseph G; Parisi, Giacomo; Wong, Bernice H; Clarke, Oliver B; Marty, Michael T; Kossiakoff, Anthony A; Khelashvili, George; Silver, David L; Mancia, Filippo
Structural basis of omega-3 fatty acid transport across the blood-brain barrier Journal Article
In: Nature, vol. 595, no. 7866, pp. 315–319, 2021, ISSN: 1476-4687.
@article{pmid34135507,
title = {Structural basis of omega-3 fatty acid transport across the blood-brain barrier},
author = {Rosemary J Cater and Geok Lin Chua and Satchal K Erramilli and James E Keener and Brendon C Choy and Piotr Tokarz and Cheen Fei Chin and Debra Q Y Quek and Brian Kloss and Joseph G Pepe and Giacomo Parisi and Bernice H Wong and Oliver B Clarke and Michael T Marty and Anthony A Kossiakoff and George Khelashvili and David L Silver and Filippo Mancia},
doi = {10.1038/s41586-021-03650-9},
issn = {1476-4687},
year = {2021},
date = {2021-07-01},
urldate = {2021-07-01},
journal = {Nature},
volume = {595},
number = {7866},
pages = {315--319},
abstract = {Docosahexaenoic acid is an omega-3 fatty acid that is essential for neurological development and function, and it is supplied to the brain and eyes predominantly from dietary sources. This nutrient is transported across the blood-brain and blood-retina barriers in the form of lysophosphatidylcholine by major facilitator superfamily domain containing 2A (MFSD2A) in a Na-dependent manner. Here we present the structure of MFSD2A determined using single-particle cryo-electron microscopy, which reveals twelve transmembrane helices that are separated into two pseudosymmetric domains. The transporter is in an inward-facing conformation and features a large amphipathic cavity that contains the Na-binding site and a bound lysolipid substrate, which we confirmed using native mass spectrometry. Together with our functional analyses and molecular dynamics simulations, this structure reveals details of how MFSD2A interacts with substrates and how Na-dependent conformational changes allow for the release of these substrates into the membrane through a lateral gate. Our work provides insights into the molecular mechanism by which this atypical major facility superfamily transporter mediates the uptake of lysolipids into the brain, and has the potential to aid in the delivery of neurotherapeutic agents.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Slezak, Tomasz; Kossiakoff, Anthony A
Engineered Ultra-High Affinity Synthetic Antibodies for SARS-CoV-2 Neutralization and Detection Journal Article
In: J Mol Biol, vol. 433, no. 10, pp. 166956, 2021, ISSN: 1089-8638.
@article{pmid33775667,
title = {Engineered Ultra-High Affinity Synthetic Antibodies for SARS-CoV-2 Neutralization and Detection},
author = {Tomasz Slezak and Anthony A Kossiakoff},
doi = {10.1016/j.jmb.2021.166956},
issn = {1089-8638},
year = {2021},
date = {2021-05-01},
urldate = {2021-05-01},
journal = {J Mol Biol},
volume = {433},
number = {10},
pages = {166956},
abstract = {The Covid-19 pandemic is a centenarial global catastrophe. Similar events are likely to be recurring with more frequency in the future. The inability to control the virus' impact is caused by many factors, but the lack of a technology infrastructure to detect and impede the virus at an early stage are principal shortcomings. Using phage display mutagenesis, we have generated a cohort of high performance antibody fragments (Fabs) that can be used in a sensitive point of care (POC) assay and are potent inhibitors (IC-0.5 nM) to viral entry into cells. The POC assay is based on a split-enzyme (β-lactamase) complementation strategy that detects virus particles at low nM levels. We have shown that this assay is equally effective for detecting other viruses like Ebola and Zika. Importantly, its components can be freeze dried and stored, but becomes fully active when rehydrated.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
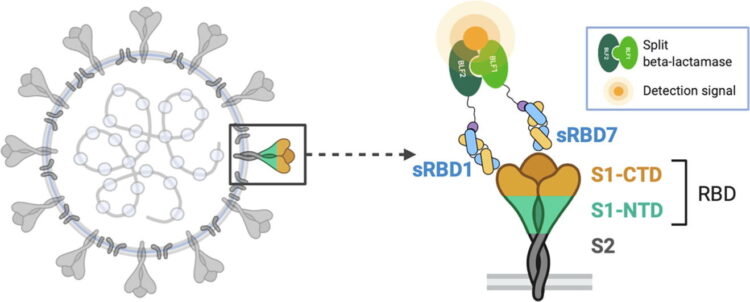
Peluso, Michael J; Takahashi, Saki; Hakim, Jill; Kelly, J Daniel; Torres, Leonel; Iyer, Nikita S; Turcios, Keirstinne; Janson, Owen; Munter, Sadie E; Thanh, Cassandra; Nixon, Christopher C; Hoh, Rebecca; Tai, Viva; Fehrman, Emily A; Hernandez, Yanel; Spinelli, Matthew A; Gandhi, Monica; Palafox, Mary-Ann; Vallari, Ana; Rodgers, Mary A; Prostko, John; Hackett, John; Trinh, Lan; Wrin, Terri; Petroplolous, Christos J; Chiu, Charles Y; Norris, Philip J; DiGermanio, Clara; Stone, Mars; Busch, Michael P; Elledge, Susanna K; Zhou, Xin X; Wells, James A; Shu, Albert; Kurtz, Theodore W; Pak, John E; Wu, Wesley; Burbelo, Peter D; Cohen, Jeffrey I; Rutishauser, Rachel L; Martin, Jeffrey N; Deeks, Steven G; Henrich, Timothy J; Rodriguez-Barraquer, Isabel; Greenhouse, Bryan
SARS-CoV-2 antibody magnitude and detectability are driven by disease severity, timing, and assay Journal Article
In: medRxiv, 2021.
@article{pmid33688675,
title = {SARS-CoV-2 antibody magnitude and detectability are driven by disease severity, timing, and assay},
author = {Michael J Peluso and Saki Takahashi and Jill Hakim and J Daniel Kelly and Leonel Torres and Nikita S Iyer and Keirstinne Turcios and Owen Janson and Sadie E Munter and Cassandra Thanh and Christopher C Nixon and Rebecca Hoh and Viva Tai and Emily A Fehrman and Yanel Hernandez and Matthew A Spinelli and Monica Gandhi and Mary-Ann Palafox and Ana Vallari and Mary A Rodgers and John Prostko and John Hackett and Lan Trinh and Terri Wrin and Christos J Petroplolous and Charles Y Chiu and Philip J Norris and Clara DiGermanio and Mars Stone and Michael P Busch and Susanna K Elledge and Xin X Zhou and James A Wells and Albert Shu and Theodore W Kurtz and John E Pak and Wesley Wu and Peter D Burbelo and Jeffrey I Cohen and Rachel L Rutishauser and Jeffrey N Martin and Steven G Deeks and Timothy J Henrich and Isabel Rodriguez-Barraquer and Bryan Greenhouse},
doi = {10.1101/2021.03.03.21251639},
year = {2021},
date = {2021-03-01},
urldate = {2021-03-01},
journal = {medRxiv},
abstract = {Serosurveillance studies are critical for estimating SARS-CoV-2 transmission and immunity, but interpretation of results is currently limited by poorly defined variability in the performance of antibody assays to detect seroreactivity over time in individuals with different clinical presentations. We measured longitudinal antibody responses to SARS-CoV-2 in plasma samples from a diverse cohort of 128 individuals over 160 days using 14 binding and neutralization assays. For all assays, we found a consistent and strong effect of disease severity on antibody magnitude, with fever, cough, hospitalization, and oxygen requirement explaining much of this variation. We found that binding assays measuring responses to spike protein had consistently higher correlation with neutralization than those measuring responses to nucleocapsid, regardless of assay format and sample timing. However, assays varied substantially with respect to sensitivity during early convalescence and in time to seroreversion. Variations in sensitivity and durability were particularly dramatic for individuals with mild infection, who had consistently lower antibody titers and represent the majority of the infected population, with sensitivities often differing substantially from reported test characteristics (e.g., amongst commercial assays, sensitivity at 6 months ranged from 33% for ARCHITECT IgG to 98% for VITROS Total Ig). Thus, the ability to detect previous infection by SARS-CoV-2 is highly dependent on the severity of the initial infection, timing relative to infection, and the assay used. These findings have important implications for the design and interpretation of SARS-CoV-2 serosurveillance studies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
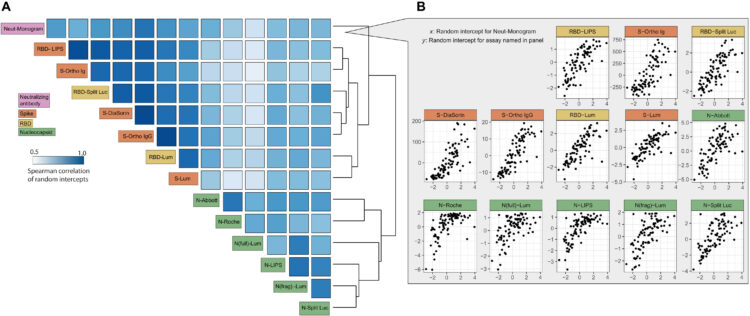
Weeks, Amy M; Byrnes, James R; Lui, Irene; Wells, James A
Mapping proteolytic neo-N termini at the surface of living cells Journal Article
In: Proc Natl Acad Sci U S A, vol. 118, no. 8, 2021, ISSN: 1091-6490.
@article{pmid33536314,
title = {Mapping proteolytic neo-N termini at the surface of living cells},
author = {Amy M Weeks and James R Byrnes and Irene Lui and James A Wells},
doi = {10.1073/pnas.2018809118},
issn = {1091-6490},
year = {2021},
date = {2021-02-01},
urldate = {2021-02-01},
journal = {Proc Natl Acad Sci U S A},
volume = {118},
number = {8},
abstract = {N terminomics is a powerful strategy for profiling proteolytic neo-N termini, but its application to cell surface proteolysis has been limited by the low relative abundance of plasma membrane proteins. Here we apply plasma membrane-targeted subtiligase variants (subtiligase-TM) to efficiently and specifically capture cell surface N termini in live cells. Using this approach, we sequenced 807 cell surface N termini and quantified changes in their abundance in response to stimuli that induce proteolytic remodeling of the cell surface proteome. To facilitate exploration of our datasets, we developed a web-accessible Atlas of Subtiligase-Captured Extracellular N Termini (ASCENT; http://wellslab.org/ascent). This technology will facilitate greater understanding of extracellular protease biology and reveal neo-N termini biomarkers and targets in disease.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
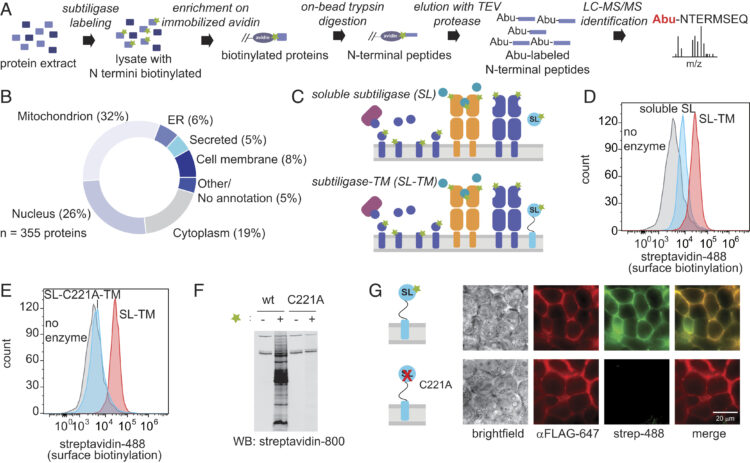
Muller, Yannick D; Ferreira, Leonardo M R; Ronin, Emilie; Ho, Patrick; Nguyen, Vinh; Faleo, Gaetano; Zhou, Yu; Lee, Karim; Leung, Kevin K; Skartsis, Nikolaos; Kaul, Anupurna M; Mulder, Arend; Claas, Frans H J; Wells, James A; Bluestone, Jeffrey A; Tang, Qizhi
Precision Engineering of an Anti-HLA-A2 Chimeric Antigen Receptor in Regulatory T Cells for Transplant Immune Tolerance Journal Article
In: Front Immunol, vol. 12, pp. 686439, 2021, ISSN: 1664-3224.
@article{pmid34616392,
title = {Precision Engineering of an Anti-HLA-A2 Chimeric Antigen Receptor in Regulatory T Cells for Transplant Immune Tolerance},
author = {Yannick D Muller and Leonardo M R Ferreira and Emilie Ronin and Patrick Ho and Vinh Nguyen and Gaetano Faleo and Yu Zhou and Karim Lee and Kevin K Leung and Nikolaos Skartsis and Anupurna M Kaul and Arend Mulder and Frans H J Claas and James A Wells and Jeffrey A Bluestone and Qizhi Tang},
doi = {10.3389/fimmu.2021.686439},
issn = {1664-3224},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Front Immunol},
volume = {12},
pages = {686439},
abstract = {Infusion of regulatory T cells (Tregs) engineered with a chimeric antigen receptor (CAR) targeting donor-derived human leukocyte antigen (HLA) is a promising strategy to promote transplant tolerance. Here, we describe an anti-HLA-A2 CAR (A2-CAR) generated by grafting the complementarity-determining regions (CDRs) of a human monoclonal anti-HLA-A2 antibody into the framework regions of the Herceptin 4D5 single-chain variable fragment and fusing it with a CD28-ζ signaling domain. The CDR-grafted A2-CAR maintained the specificity of the original antibody. We then generated HLA-A2 mono-specific human CAR Tregs either by deleting the endogenous T-cell receptor (TCR) CRISPR/Cas9 and introducing the A2-CAR using lentiviral transduction or by directly integrating the CAR construct into the TCR alpha constant locus using homology-directed repair. These A2-CARTCR human Tregs maintained both Treg phenotype and function . Moreover, they selectively accumulated in HLA-A2-expressing islets transplanted from either HLA-A2 transgenic mice or deceased human donors. A2-CARTCR Tregs did not impair the function of these HLA-A2 islets, whereas similarly engineered A2-CARTCRCD4 conventional T cells rejected the islets in less than 2 weeks. A2-CARTCR Tregs delayed graft--host disease only in the presence of HLA-A2, expressed either by co-transferred peripheral blood mononuclear cells or by the recipient mice. Altogether, we demonstrate that genome-engineered mono-antigen-specific A2-CAR Tregs localize to HLA-A2-expressing grafts and exhibit antigen-dependent suppression, independent of TCR expression. These approaches may be applied towards developing precision Treg cell therapies for transplant tolerance.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
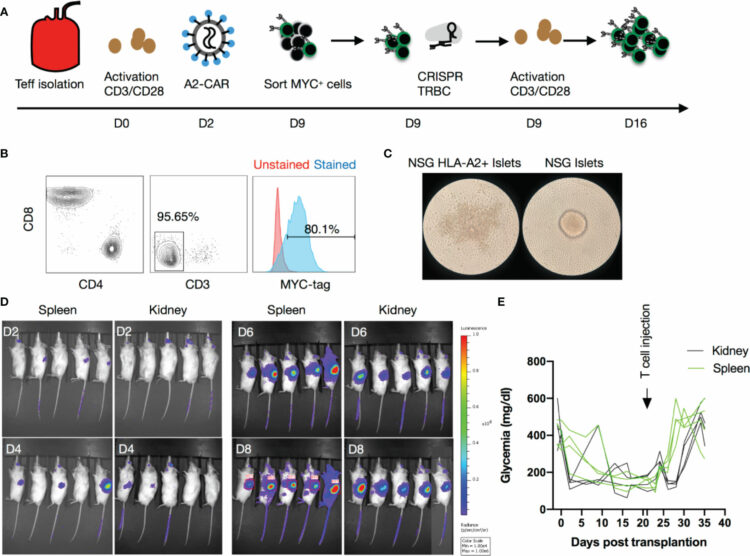
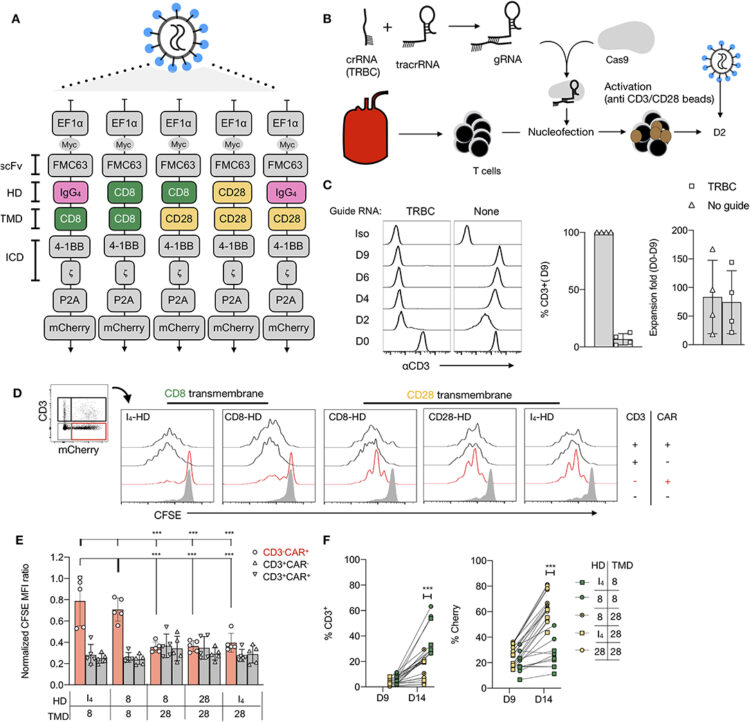
Muller, Yannick D; Nguyen, Duy P; Ferreira, Leonardo M R; Ho, Patrick; Raffin, Caroline; Valencia, Roxxana Valeria Beltran; Congrave-Wilson, Zion; Roth, Theodore L; Eyquem, Justin; Gool, Frederic Van; Marson, Alexander; Perez, Laurent; Wells, James A; Bluestone, Jeffrey A; Tang, Qizhi
The CD28-Transmembrane Domain Mediates Chimeric Antigen Receptor Heterodimerization With CD28 Journal Article
In: Front Immunol, vol. 12, pp. 639818, 2021, ISSN: 1664-3224.
@article{pmid33833759,
title = {The CD28-Transmembrane Domain Mediates Chimeric Antigen Receptor Heterodimerization With CD28},
author = {Yannick D Muller and Duy P Nguyen and Leonardo M R Ferreira and Patrick Ho and Caroline Raffin and Roxxana Valeria Beltran Valencia and Zion Congrave-Wilson and Theodore L Roth and Justin Eyquem and Frederic Van Gool and Alexander Marson and Laurent Perez and James A Wells and Jeffrey A Bluestone and Qizhi Tang},
doi = {10.3389/fimmu.2021.639818},
issn = {1664-3224},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Front Immunol},
volume = {12},
pages = {639818},
abstract = {Anti-CD19 chimeric antigen receptor (CD19-CAR)-engineered T cells are approved therapeutics for malignancies. The impact of the hinge domain (HD) and the transmembrane domain (TMD) between the extracellular antigen-targeting CARs and the intracellular signaling modalities of CARs has not been systemically studied. In this study, a series of 19-CARs differing only by their HD (CD8, CD28, or IgG) and TMD (CD8 or CD28) was generated. CARs containing a CD28-TMD, but not a CD8-TMD, formed heterodimers with the endogenous CD28 in human T cells, as shown by co-immunoprecipitation and CAR-dependent proliferation of anti-CD28 stimulation. This dimerization was dependent on polar amino acids in the CD28-TMD and was more efficient with CARs containing CD28 or CD8 HD than IgG-HD. The CD28-CAR heterodimers did not respond to CD80 and CD86 stimulation but had a significantly reduced CD28 cell-surface expression. These data unveiled a fundamental difference between CD28-TMD and CD8-TMD and indicated that CD28-TMD can modulate CAR T-cell activities by engaging endogenous partners.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
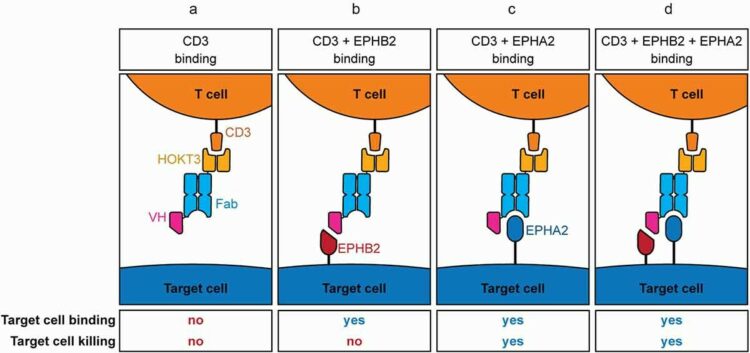
Enderle, Leonie; Shalaby, Karim H; Gorelik, Maryna; Weiss, Alexander; Blazer, Levi L; Paduch, Marcin; Cardarelli, Lia; Kossiakoff, Anthony; Adams, Jarrett J; Sidhu, Sachdev S
A T cell redirection platform for co-targeting dual antigens on solid tumors Journal Article
In: MAbs, vol. 13, no. 1, pp. 1933690, 2021, ISSN: 1942-0870.
@article{pmid34190031,
title = {A T cell redirection platform for co-targeting dual antigens on solid tumors},
author = {Leonie Enderle and Karim H Shalaby and Maryna Gorelik and Alexander Weiss and Levi L Blazer and Marcin Paduch and Lia Cardarelli and Anthony Kossiakoff and Jarrett J Adams and Sachdev S Sidhu},
doi = {10.1080/19420862.2021.1933690},
issn = {1942-0870},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {MAbs},
volume = {13},
number = {1},
pages = {1933690},
abstract = {In order to direct T cells to specific features of solid cancer cells, we engineered a bispecific antibody format, named ual ntigen cell ngager (DATE), by fusing a single-chain variable fragment targeting CD3 to a tumor-targeting antigen-binding fragment. In this format, multiple novel paratopes against different tumor antigens were able to recruit T-cell cytotoxicity to tumor cells and in an pancreatic ductal adenocarcinoma xenograft model. Since unique surface antigens in solid tumors are limited, in order to enhance selectivity, we further engineered "double-DATEs" targeting two tumor antigens simultaneously. The double-DATE contains an additional autonomous variable heavy-chain domain, which binds a second tumor antigen without itself eliciting a cytotoxic response. This novel modality provides a strategy to enhance the selectivity of immune redirection through binary targeting of native tumor antigens. The modularity and use of a common, stable human framework for all components enables a pipeline approach to rapidly develop a broad repertoire of tailored DATEs and double-DATEs with favorable biophysical properties and high potencies and selectivities.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
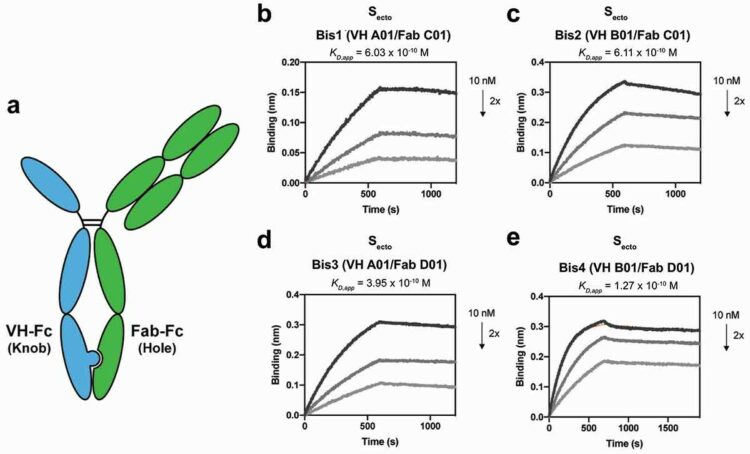
Lim, Shion A; Gramespacher, Josef A; Pance, Katarina; Rettko, Nicholas J; Solomon, Paige; Jin, Jing; Lui, Irene; Elledge, Susanna K; Liu, Jia; Bracken, Colton J; Simmons, Graham; Zhou, Xin X; Leung, Kevin K; Wells, James A
Bispecific VH/Fab antibodies targeting neutralizing and non-neutralizing Spike epitopes demonstrate enhanced potency against SARS-CoV-2 Journal Article
In: MAbs, vol. 13, no. 1, pp. 1893426, 2021, ISSN: 1942-0870.
@article{pmid33666135,
title = {Bispecific VH/Fab antibodies targeting neutralizing and non-neutralizing Spike epitopes demonstrate enhanced potency against SARS-CoV-2},
author = {Shion A Lim and Josef A Gramespacher and Katarina Pance and Nicholas J Rettko and Paige Solomon and Jing Jin and Irene Lui and Susanna K Elledge and Jia Liu and Colton J Bracken and Graham Simmons and Xin X Zhou and Kevin K Leung and James A Wells},
doi = {10.1080/19420862.2021.1893426},
issn = {1942-0870},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {MAbs},
volume = {13},
number = {1},
pages = {1893426},
abstract = {Numerous neutralizing antibodies that target SARS-CoV-2 have been reported, and most directly block binding of the viral Spike receptor-binding domain (RBD) to angiotensin-converting enzyme II (ACE2). Here, we deliberately exploit non-neutralizing RBD antibodies, showing they can dramatically assist in neutralization when linked to neutralizing binders. We identified antigen-binding fragments (Fabs) by phage display that bind RBD, but do not block ACE2 or neutralize virus as IgGs. When these non-neutralizing Fabs were assembled into bispecific VH/Fab IgGs with a neutralizing VH domain, we observed a ~ 25-fold potency improvement in neutralizing SARS-CoV-2 compared to the mono-specific bi-valent VH-Fc alone or the cocktail of the VH-Fc and IgG. This effect was epitope-dependent, reflecting the unique geometry of the bispecific antibody toward Spike. Our results show that a bispecific antibody that combines both neutralizing and non-neutralizing epitopes on Spike-RBD is a promising and rapid engineering strategy to improve the potency of SARS-CoV-2 antibodies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
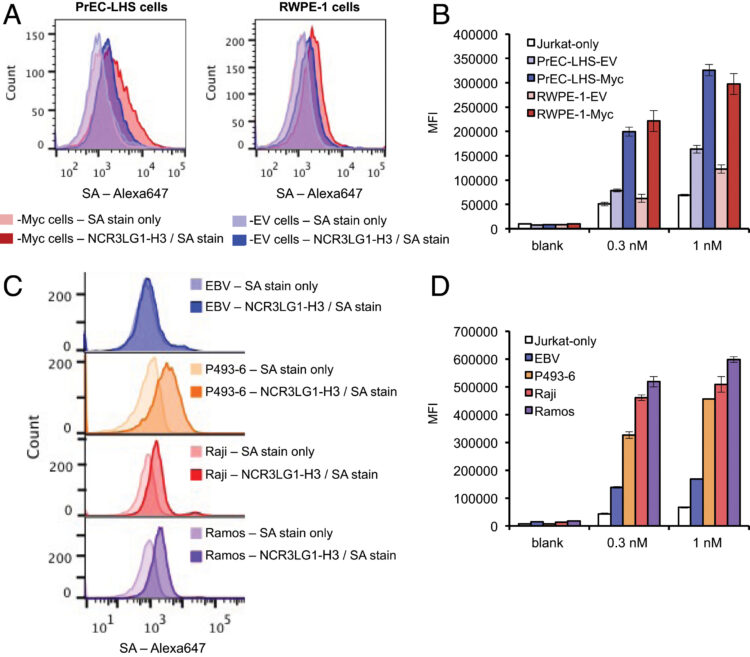
Chen, Wentao; Mou, Kurt Yun; Solomon, Paige; Aggarwal, Rahul; Leung, Kevin K; Wells, James A
Large remodeling of the Myc-induced cell surface proteome in B cells and prostate cells creates new opportunities for immunotherapy Journal Article
In: Proc Natl Acad Sci U S A, vol. 118, no. 4, 2021, ISSN: 1091-6490.
@article{pmid33483421,
title = {Large remodeling of the Myc-induced cell surface proteome in B cells and prostate cells creates new opportunities for immunotherapy},
author = {Wentao Chen and Kurt Yun Mou and Paige Solomon and Rahul Aggarwal and Kevin K Leung and James A Wells},
doi = {10.1073/pnas.2018861118},
issn = {1091-6490},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Proc Natl Acad Sci U S A},
volume = {118},
number = {4},
abstract = {MYC is a powerful transcription factor overexpressed in many human cancers including B cell and prostate cancers. Antibody therapeutics are exciting opportunities to attack cancers but require knowledge of surface proteins that change due to oncogene expression. To identify how MYC overexpression remodels the cell surface proteome in a cell autologous fashion and in different cell types, we investigated the impact of MYC overexpression on 800 surface proteins in three isogenic model cell lines either of B cell or prostate cell origin engineered to have high or low MYC levels. We found that MYC overexpression resulted in dramatic remodeling (both up- and down-regulation) of the cell surfaceome in a cell type-dependent fashion. We found systematic and large increases in distinct sets of >80 transporters including nucleoside transporters and nutrient transporters making cells more sensitive to toxic nucleoside analogs like cytarabine, commonly used for treating hematological cancers. Paradoxically, MYC overexpression also increased expression of surface proteins driving cell turnover such as TNFRSF10B, also known as death receptor 5, and immune cell attacking signals such as the natural killer cell activating ligand NCR3LG1, also known as B7-H6. We generated recombinant antibodies to these two targets and verified their up-regulation in MYC overexpression cell lines and showed they were sensitive to bispecific T cell engagers (BiTEs). Our studies demonstrate how MYC overexpression leads to dramatic bidirectional remodeling of the surfaceome in a cell type-dependent but functionally convergent fashion and identify surface targets or combinations thereof as possible candidates for cytotoxic metabolite or immunotherapy.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
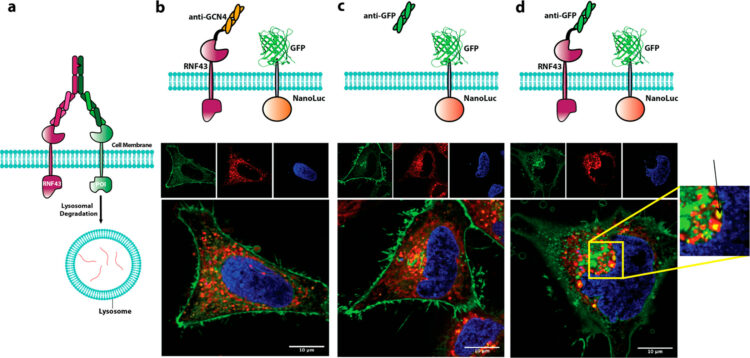
Cotton, Adam D; Nguyen, Duy P; Gramespacher, Josef A; Seiple, Ian B; Wells, James A
Development of Antibody-Based PROTACs for the Degradation of the Cell-Surface Immune Checkpoint Protein PD-L1 Journal Article
In: J Am Chem Soc, vol. 143, no. 2, pp. 593–598, 2021, ISSN: 1520-5126.
@article{pmid33395526,
title = {Development of Antibody-Based PROTACs for the Degradation of the Cell-Surface Immune Checkpoint Protein PD-L1},
author = {Adam D Cotton and Duy P Nguyen and Josef A Gramespacher and Ian B Seiple and James A Wells},
doi = {10.1021/jacs.0c10008},
issn = {1520-5126},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {J Am Chem Soc},
volume = {143},
number = {2},
pages = {593--598},
abstract = {Targeted protein degradation has emerged as a new paradigm to manipulate cellular proteostasis. Proteolysis-targeting chimeras (PROTACs) are bifunctional small molecules that recruit an E3 ligase to a target protein of interest, promoting its ubiquitination and subsequent degradation. Here, we report the development of antibody-based PROTACs (AbTACs), fully recombinant bispecific antibodies that recruit membrane-bound E3 ligases for the degradation of cell-surface proteins. We show that an AbTAC can induce the lysosomal degradation of programmed death-ligand 1 by recruitment of the membrane-bound E3 ligase RNF43. AbTACs represent a new archetype within the PROTAC field to target cell-surface proteins with fully recombinant biological molecules.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}