Publications: 2020
2020
Glasgow, Anum; Glasgow, Jeff; Limonta, Daniel; Solomon, Paige; Lui, Irene; Zhang, Yang; Nix, Matthew A; Rettko, Nicholas J; Zha, Shoshana; Yamin, Rachel; Kao, Kevin; Rosenberg, Oren S; Ravetch, Jeffrey V; Wiita, Arun P; Leung, Kevin K; Lim, Shion A; Zhou, Xin X; Hobman, Tom C; Kortemme, Tanja; Wells, James A
Engineered ACE2 receptor traps potently neutralize SARS-CoV-2 Journal Article
In: Proc Natl Acad Sci U S A, vol. 117, no. 45, pp. 28046–28055, 2020, ISSN: 1091-6490.
@article{pmid33093202,
title = {Engineered ACE2 receptor traps potently neutralize SARS-CoV-2},
author = {Anum Glasgow and Jeff Glasgow and Daniel Limonta and Paige Solomon and Irene Lui and Yang Zhang and Matthew A Nix and Nicholas J Rettko and Shoshana Zha and Rachel Yamin and Kevin Kao and Oren S Rosenberg and Jeffrey V Ravetch and Arun P Wiita and Kevin K Leung and Shion A Lim and Xin X Zhou and Tom C Hobman and Tanja Kortemme and James A Wells},
doi = {10.1073/pnas.2016093117},
issn = {1091-6490},
year = {2020},
date = {2020-11-01},
urldate = {2020-11-01},
journal = {Proc Natl Acad Sci U S A},
volume = {117},
number = {45},
pages = {28046--28055},
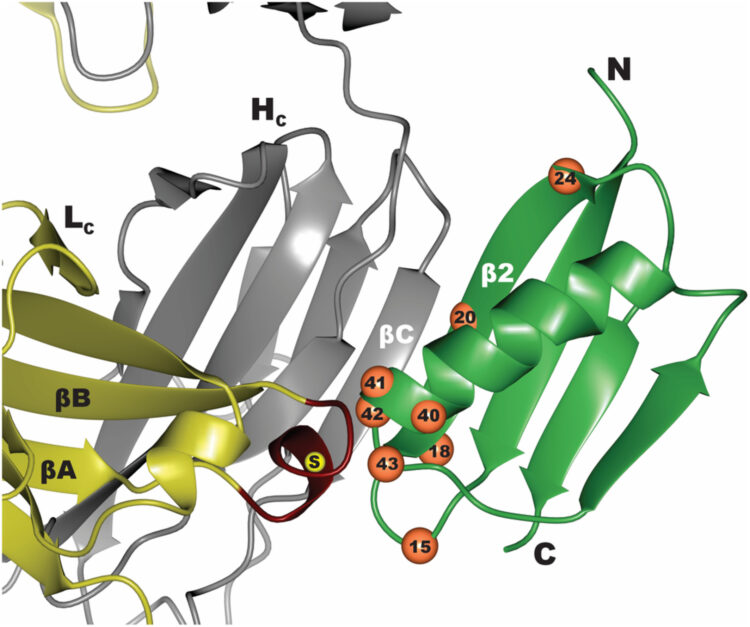
abstract = {An essential mechanism for severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection begins with the viral spike protein binding to the human receptor protein angiotensin-converting enzyme II (ACE2). Here, we describe a stepwise engineering approach to generate a set of affinity optimized, enzymatically inactivated ACE2 variants that potently block SARS-CoV-2 infection of cells. These optimized receptor traps tightly bind the receptor binding domain (RBD) of the viral spike protein and prevent entry into host cells. We first computationally designed the ACE2-RBD interface using a two-stage flexible protein backbone design process that improved affinity for the RBD by up to 12-fold. These designed receptor variants were affinity matured an additional 14-fold by random mutagenesis and selection using yeast surface display. The highest-affinity variant contained seven amino acid changes and bound to the RBD 170-fold more tightly than wild-type ACE2. With the addition of the natural ACE2 collectrin domain and fusion to a human immunoglobulin crystallizable fragment (Fc) domain for increased stabilization and avidity, the most optimal ACE2 receptor traps neutralized SARS-CoV-2-pseudotyped lentivirus and authentic SARS-CoV-2 virus with half-maximal inhibitory concentrations (IC50s) in the 10- to 100-ng/mL range. Engineered ACE2 receptor traps offer a promising route to fighting infections by SARS-CoV-2 and other ACE2-using coronaviruses, with the key advantage that viral resistance would also likely impair viral entry. Moreover, such traps can be predesigned for viruses with known entry receptors for faster therapeutic response without the need for neutralizing antibodies isolated from convalescent patients.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Zamecnik, Colin R; Rajan, Jayant V; Yamauchi, Kevin A; Mann, Sabrina A; Loudermilk, Rita P; Sowa, Gavin M; Zorn, Kelsey C; Alvarenga, Bonny D; Gaebler, Christian; Caskey, Marina; Stone, Mars; Norris, Philip J; Gu, Wei; Chiu, Charles Y; Ng, Dianna; Byrnes, James R; Zhou, Xin X; Wells, James A; Robbiani, Davide F; Nussenzweig, Michel C; DeRisi, Joseph L; Wilson, Michael R
ReScan, a Multiplex Diagnostic Pipeline, Pans Human Sera for SARS-CoV-2 Antigens Journal Article
In: Cell Rep Med, vol. 1, no. 7, pp. 100123, 2020, ISSN: 2666-3791.
@article{pmid32995758,
title = {ReScan, a Multiplex Diagnostic Pipeline, Pans Human Sera for SARS-CoV-2 Antigens},
author = {Colin R Zamecnik and Jayant V Rajan and Kevin A Yamauchi and Sabrina A Mann and Rita P Loudermilk and Gavin M Sowa and Kelsey C Zorn and Bonny D Alvarenga and Christian Gaebler and Marina Caskey and Mars Stone and Philip J Norris and Wei Gu and Charles Y Chiu and Dianna Ng and James R Byrnes and Xin X Zhou and James A Wells and Davide F Robbiani and Michel C Nussenzweig and Joseph L DeRisi and Michael R Wilson},
doi = {10.1016/j.xcrm.2020.100123},
issn = {2666-3791},
year = {2020},
date = {2020-10-01},
urldate = {2020-10-01},
journal = {Cell Rep Med},
volume = {1},
number = {7},
pages = {100123},
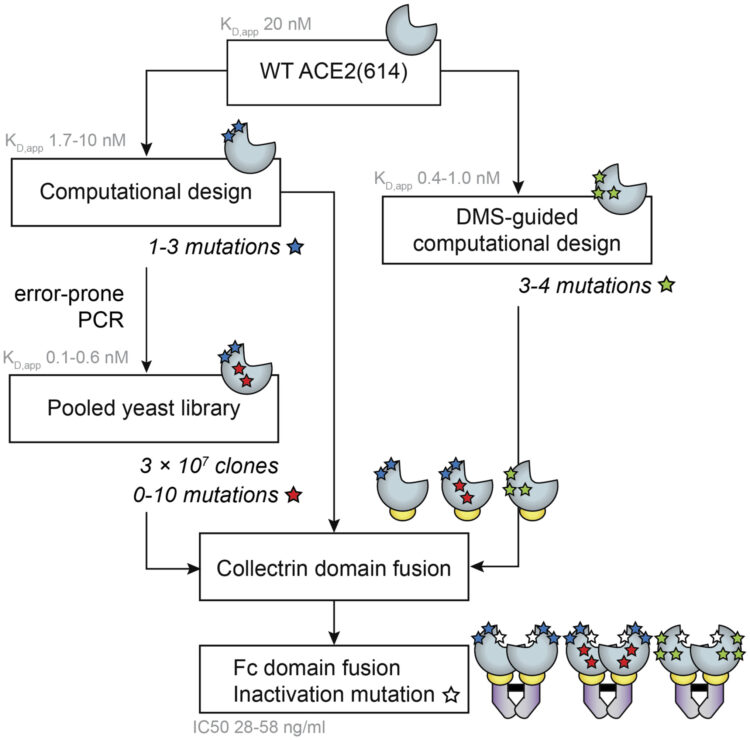
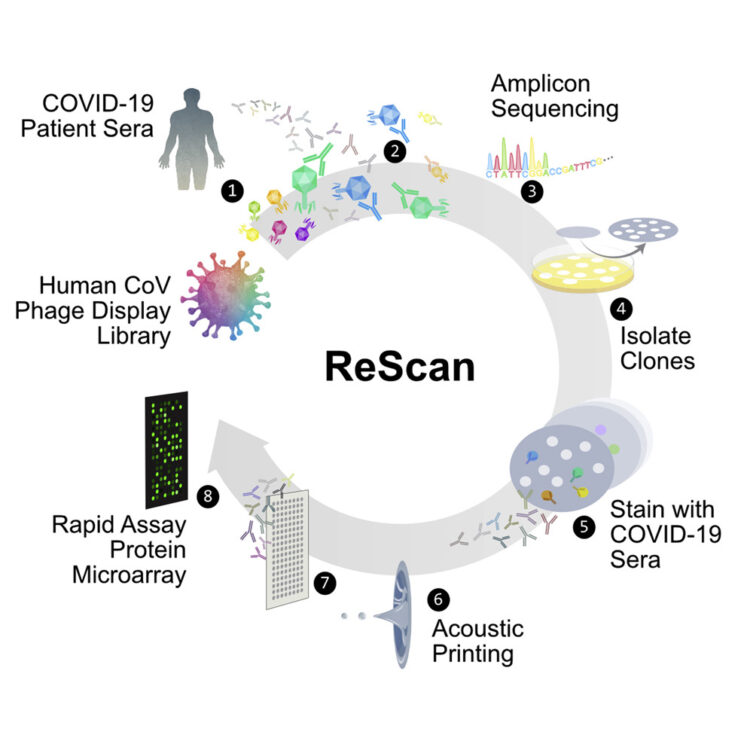
abstract = {Comprehensive understanding of the serological response to SARS-CoV-2 infection is important for both pathophysiologic insight and diagnostic development. Here, we generate a pan-human coronavirus programmable phage display assay to perform proteome-wide profiling of coronavirus antigens enriched by 98 COVID-19 patient sera. Next, we use ReScan, a method to efficiently sequester phage expressing the most immunogenic peptides and print them onto paper-based microarrays using acoustic liquid handling, which isolates and identifies nine candidate antigens, eight of which are derived from the two proteins used for SARS-CoV-2 serologic assays: spike and nucleocapsid proteins. After deployment in a high-throughput assay amenable to clinical lab settings, these antigens show improved specificity over a whole protein panel. This proof-of-concept study demonstrates that ReScan will have broad applicability for other emerging infectious diseases or autoimmune diseases that lack a valid biomarker, enabling a seamless pipeline from antigen discovery to diagnostic using one recombinant protein source.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Mahuron, Kelly M.; Moreau, Joshua M.; Glasgow, Jeff E.; Boda, Devi P.; Pauli, Mariela L.; Gouirand, Victoire; Panjabi, Luv; Grewal, Robby; Luber, Jacob M.; Mathur, Anubhav N.; Feldman, Renny M.; Shifrut, Eric; Mehta, Pooja; Lowe, Margaret M.; Alvarado, Michael D.; Marson, Alexander; Singer, Meromit; Wells, Jim; Jupp, Ray; Daud, Adil I.; Rosenblum, Michael D.
Layilin augments integrin activation to promote antitumor immunity Journal Article
In: vol. 217, no. 9, 2020, ISSN: 1540-9538.
@article{Mahuron2020,
title = {Layilin augments integrin activation to promote antitumor immunity},
author = {Kelly M. Mahuron and Joshua M. Moreau and Jeff E. Glasgow and Devi P. Boda and Mariela L. Pauli and Victoire Gouirand and Luv Panjabi and Robby Grewal and Jacob M. Luber and Anubhav N. Mathur and Renny M. Feldman and Eric Shifrut and Pooja Mehta and Margaret M. Lowe and Michael D. Alvarado and Alexander Marson and Meromit Singer and Jim Wells and Ray Jupp and Adil I. Daud and Michael D. Rosenblum},
doi = {10.1084/jem.20192080},
issn = {1540-9538},
year = {2020},
date = {2020-09-07},
urldate = {2020-09-07},
volume = {217},
number = {9},
publisher = {Rockefeller University Press},
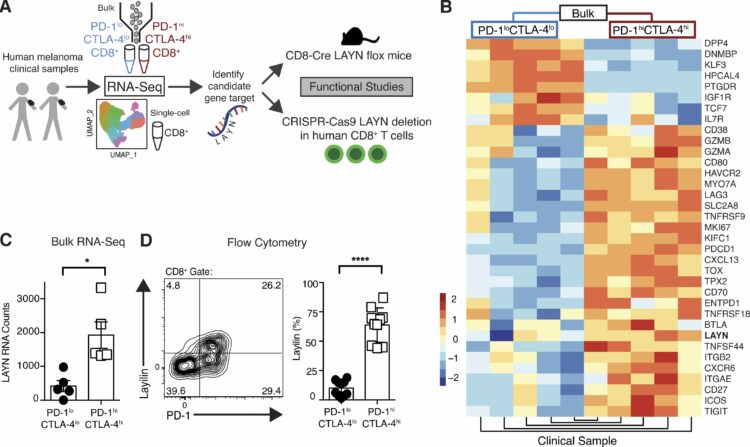
abstract = {<jats:p>Tumor-infiltrating CD8+ T cells mediate antitumor immune responses. However, the mechanisms by which T cells remain poised to kill cancer cells despite expressing high levels of inhibitory receptors are unknown. Here, we report that layilin, a C-type lectin domain–containing membrane glycoprotein, is selectively expressed on highly activated, clonally expanded, but phenotypically exhausted CD8+ T cells in human melanoma. Lineage-specific deletion of layilin on murine CD8+ T cells reduced their accumulation in tumors and increased tumor growth in vivo. Congruently, gene editing of LAYN in human CD8+ T cells reduced direct tumor cell killing ex vivo. On a molecular level, layilin colocalized with integrin αLβ2 (LFA-1) on T cells, and cross-linking layilin promoted the activated state of this integrin. Accordingly, LAYN deletion resulted in attenuated LFA-1–dependent cellular adhesion. Collectively, our results identify layilin as part of a molecular pathway in which exhausted or “dysfunctional” CD8+ T cells enhance cellular adhesiveness to maintain their cytotoxic potential.</jats:p>},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
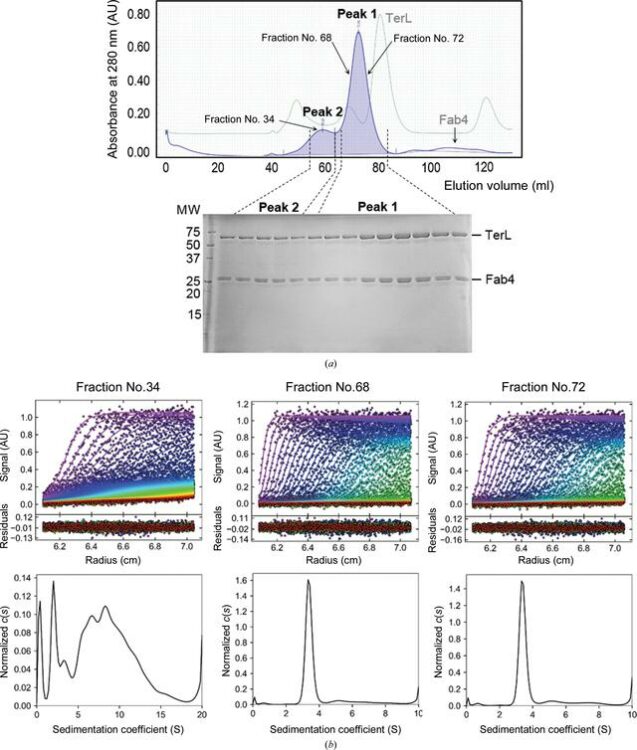
Lokareddy, Ravi K; Ko, Ying Hui; Hong, Nathaniel; Doll, Steven G; Paduch, Marcin; Niederweis, Michael; Kossiakoff, Anthony A; Cingolani, Gino
Recognition of an α-helical hairpin in P22 large terminase by a synthetic antibody fragment Journal Article
In: Acta Crystallogr D Struct Biol, vol. 76, no. Pt 9, pp. 876–888, 2020, ISSN: 2059-7983.
@article{pmid32876063,
title = {Recognition of an α-helical hairpin in P22 large terminase by a synthetic antibody fragment},
author = {Ravi K Lokareddy and Ying Hui Ko and Nathaniel Hong and Steven G Doll and Marcin Paduch and Michael Niederweis and Anthony A Kossiakoff and Gino Cingolani},
doi = {10.1107/S2059798320009912},
issn = {2059-7983},
year = {2020},
date = {2020-09-01},
urldate = {2020-09-01},
journal = {Acta Crystallogr D Struct Biol},
volume = {76},
number = {Pt 9},
pages = {876--888},
abstract = {The genome-packaging motor of tailed bacteriophages and herpesviruses is a multisubunit protein complex formed by several copies of a large (TerL) and a small (TerS) terminase subunit. The motor assembles transiently at the portal protein vertex of an empty precursor capsid to power the energy-dependent packaging of viral DNA. Both the ATPase and nuclease activities associated with genome packaging reside in TerL. Structural studies of TerL from bacteriophage P22 have been hindered by the conformational flexibility of this enzyme and its susceptibility to proteolysis. Here, an unbiased, synthetic phage-display Fab library was screened and a panel of high-affinity Fabs against P22 TerL were identified. This led to the discovery of a recombinant antibody fragment, Fab4, that binds a 33-amino-acid α-helical hairpin at the N-terminus of TerL with an equilibrium dissociation constant K of 71.5 nM. A 1.51 Å resolution crystal structure of Fab4 bound to the TerL epitope (TLE) together with a 1.15 Å resolution crystal structure of the unliganded Fab4, which is the highest resolution ever achieved for a Fab, elucidate the principles governing the recognition of this novel helical epitope. TLE adopts two different conformations in the asymmetric unit and buries as much as 1250 Å of solvent-accessible surface in Fab4. TLE recognition is primarily mediated by conformational changes in the third complementarity-determining region of the Fab4 heavy chain (CDR H3) that take place upon epitope binding. It is demonstrated that TLE can be introduced genetically at the N-terminus of a target protein, where it retains high-affinity binding to Fab4.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
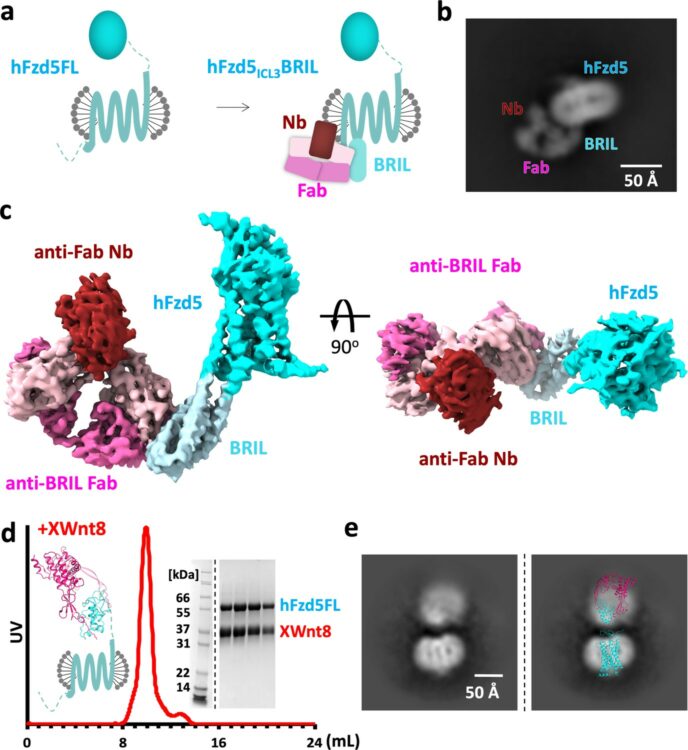
Tsutsumi, Naotaka; Mukherjee, Somnath; Waghray, Deepa; Janda, Claudia Y; Jude, Kevin M; Miao, Yi; Burg, John S; Aduri, Nanda Gowtham; Kossiakoff, Anthony A; Gati, Cornelius; Garcia, K Christopher
Structure of human Frizzled5 by fiducial-assisted cryo-EM supports a heterodimeric mechanism of canonical Wnt signaling Journal Article
In: Elife, vol. 9, 2020, ISSN: 2050-084X.
@article{pmid32762848,
title = {Structure of human Frizzled5 by fiducial-assisted cryo-EM supports a heterodimeric mechanism of canonical Wnt signaling},
author = {Naotaka Tsutsumi and Somnath Mukherjee and Deepa Waghray and Claudia Y Janda and Kevin M Jude and Yi Miao and John S Burg and Nanda Gowtham Aduri and Anthony A Kossiakoff and Cornelius Gati and K Christopher Garcia},
doi = {10.7554/eLife.58464},
issn = {2050-084X},
year = {2020},
date = {2020-08-01},
urldate = {2020-08-01},
journal = {Elife},
volume = {9},
abstract = {Frizzleds (Fzd) are the primary receptors for Wnt morphogens, which are essential regulators of stem cell biology, yet the structural basis of Wnt signaling through Fzd remains poorly understood. Here we report the structure of an unliganded human Fzd5 determined by single-particle cryo-EM at 3.7 Å resolution, with the aid of an antibody chaperone acting as a fiducial marker. We also analyzed the topology of low-resolution XWnt8/Fzd5 complex particles, which revealed extreme flexibility between the Wnt/Fzd-CRD and the Fzd-TM regions. Analysis of Wnt/β-catenin signaling in response to Wnt3a versus a 'surrogate agonist' that cross-links Fzd to LRP6, revealed identical structure-activity relationships. Thus, canonical Wnt/β-catenin signaling appears to be principally reliant on ligand-induced Fzd/LRP6 heterodimerization, versus the allosteric mechanisms seen in structurally analogous class A G protein-coupled receptors, and Smoothened. These findings deepen our mechanistic understanding of Wnt signal transduction, and have implications for harnessing Wnt agonism in regenerative medicine.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
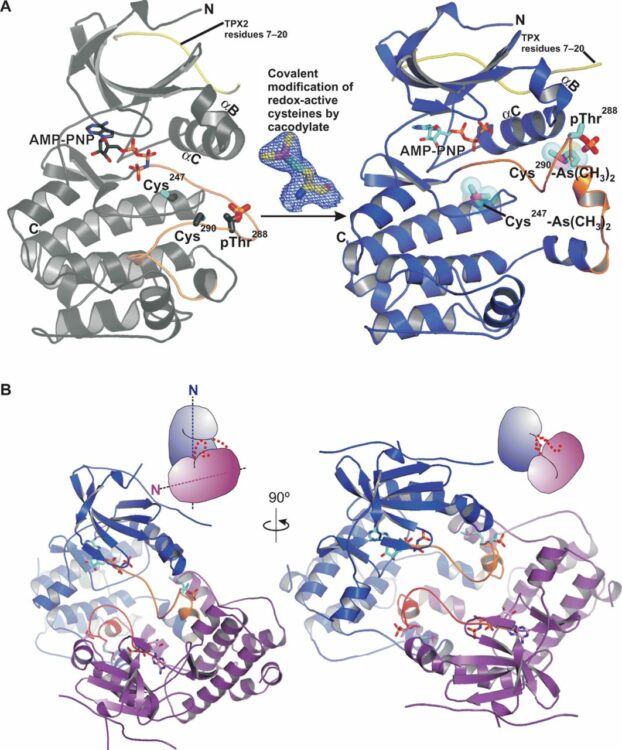
Lim, Daniel C.; Joukov, Vladimir; Rettenmaier, T. Justin; Kumagai, Akiko; Dunphy, William G.; Wells, James A.; Yaffe, Michael B.
Redox priming promotes Aurora A activation during mitosis Journal Article
In: Sci. Signal., vol. 13, no. 641, 2020, ISSN: 1937-9145.
@article{Lim2020,
title = {Redox priming promotes Aurora A activation during mitosis},
author = {Daniel C. Lim and Vladimir Joukov and T. Justin Rettenmaier and Akiko Kumagai and William G. Dunphy and James A. Wells and Michael B. Yaffe},
doi = {10.1126/scisignal.abb6707},
issn = {1937-9145},
year = {2020},
date = {2020-07-21},
urldate = {2020-07-21},
journal = {Sci. Signal.},
volume = {13},
number = {641},
publisher = {American Association for the Advancement of Science (AAAS)},
abstract = {<jats:p>CoAlation of the kinase Aurora A induces an open dimer conformation that facilitates its activation.</jats:p>},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
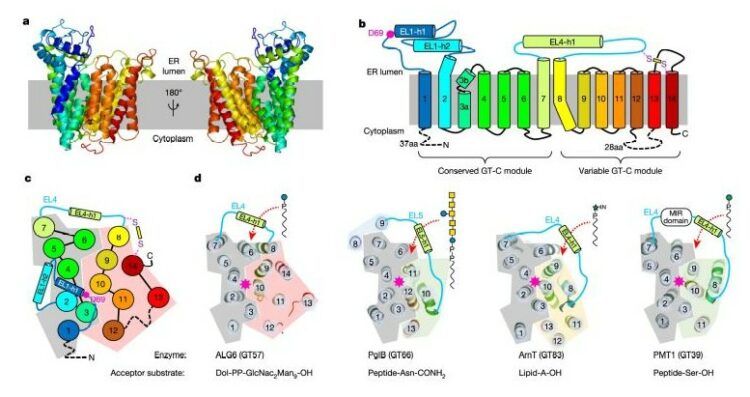
Bloch, Joël S; Pesciullesi, Giorgio; Boilevin, Jérémy; Nosol, Kamil; Irobalieva, Rossitza N; Darbre, Tamis; Aebi, Markus; Kossiakoff, Anthony A; Reymond, Jean-Louis; Locher, Kaspar P
Structure and mechanism of the ER-based glucosyltransferase ALG6 Journal Article
In: Nature, vol. 579, no. 7799, pp. 443–447, 2020, ISSN: 1476-4687.
@article{pmid32103179,
title = {Structure and mechanism of the ER-based glucosyltransferase ALG6},
author = {Joël S Bloch and Giorgio Pesciullesi and Jérémy Boilevin and Kamil Nosol and Rossitza N Irobalieva and Tamis Darbre and Markus Aebi and Anthony A Kossiakoff and Jean-Louis Reymond and Kaspar P Locher},
doi = {10.1038/s41586-020-2044-z},
issn = {1476-4687},
year = {2020},
date = {2020-03-01},
urldate = {2020-03-01},
journal = {Nature},
volume = {579},
number = {7799},
pages = {443--447},
abstract = {In eukaryotic protein N-glycosylation, a series of glycosyltransferases catalyse the biosynthesis of a dolichylpyrophosphate-linked oligosaccharide before its transfer onto acceptor proteins. The final seven steps occur in the lumen of the endoplasmic reticulum (ER) and require dolichylphosphate-activated mannose and glucose as donor substrates. The responsible enzymes-ALG3, ALG9, ALG12, ALG6, ALG8 and ALG10-are glycosyltransferases of the C-superfamily (GT-Cs), which are loosely defined as containing membrane-spanning helices and processing an isoprenoid-linked carbohydrate donor substrate. Here we present the cryo-electron microscopy structure of yeast ALG6 at 3.0 Å resolution, which reveals a previously undescribed transmembrane protein fold. Comparison with reported GT-C structures suggests that GT-C enzymes contain a modular architecture with a conserved module and a variable module, each with distinct functional roles. We used synthetic analogues of dolichylphosphate-linked and dolichylpyrophosphate-linked sugars and enzymatic glycan extension to generate donor and acceptor substrates using purified enzymes of the ALG pathway to recapitulate the activity of ALG6 in vitro. A second cryo-electron microscopy structure of ALG6 bound to an analogue of dolichylphosphate-glucose at 3.9 Å resolution revealed the active site of the enzyme. Functional analysis of ALG6 variants identified a catalytic aspartate residue that probably acts as a general base. This residue is conserved in the GT-C superfamily. Our results define the architecture of ER-luminal GT-C enzymes and provide a structural basis for understanding their catalytic mechanisms.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Mukherjee, Somnath; Erramilli, Satchal K; Ammirati, Mark; Alvarez, Frances J D; Fennell, Kimberly F; Purdy, Michael D; Skrobek, Blazej M; Radziwon, Katarzyna; Coukos, John; Kang, Yanyong; Dutka, Przemysław; Gao, Xiang; Qiu, Xiayang; Yeager, Mark; Xu, H Eric; Han, Seungil; Kossiakoff, Anthony A
Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins Journal Article
In: Nat Commun, vol. 11, no. 1, pp. 1598, 2020, ISSN: 2041-1723.
@article{pmid32221310,
title = {Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins},
author = {Somnath Mukherjee and Satchal K Erramilli and Mark Ammirati and Frances J D Alvarez and Kimberly F Fennell and Michael D Purdy and Blazej M Skrobek and Katarzyna Radziwon and John Coukos and Yanyong Kang and Przemysław Dutka and Xiang Gao and Xiayang Qiu and Mark Yeager and H Eric Xu and Seungil Han and Anthony A Kossiakoff},
doi = {10.1038/s41467-020-15363-0},
issn = {2041-1723},
year = {2020},
date = {2020-03-01},
urldate = {2020-03-01},
journal = {Nat Commun},
volume = {11},
number = {1},
pages = {1598},
abstract = {We propose the concept of universal fiducials based on a set of pre-made semi-synthetic antibodies (sABs) generated by customized phage display selections against the fusion protein BRIL, an engineered variant of apocytochrome b562a. These sABs can bind to BRIL fused either into the loops or termini of different GPCRs, ion channels, receptors and transporters without disrupting their structure. A crystal structure of BRIL in complex with an affinity-matured sAB (BAG2) that bound to all systems tested delineates the footprint of interaction. Negative stain and cryoEM data of several examples of BRIL-membrane protein chimera highlight the effectiveness of the sABs as universal fiducial marks. Taken together with a cryoEM structure of sAB bound human nicotinic acetylcholine receptor, this work demonstrates that these anti-BRIL sABs can greatly enhance the particle properties leading to improved cryoEM outcomes, especially for challenging membrane proteins.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
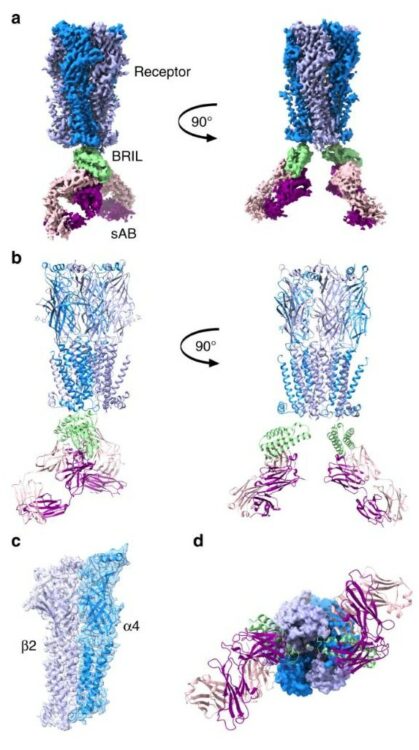
Slezak, Tomasz; Bailey, Lucas J; Jaskolowski, Mateusz; Nahotko, Dominik A; Filippova, Ekaterina V; Davydova, Elena K; Kossiakoff, Anthony A
An engineered ultra-high affinity Fab-Protein G pair enables a modular antibody platform with multifunctional capability Journal Article
In: Protein Sci, vol. 29, no. 1, pp. 141–156, 2020, ISSN: 1469-896X.
@article{pmid31622515,
title = {An engineered ultra-high affinity Fab-Protein G pair enables a modular antibody platform with multifunctional capability},
author = {Tomasz Slezak and Lucas J Bailey and Mateusz Jaskolowski and Dominik A Nahotko and Ekaterina V Filippova and Elena K Davydova and Anthony A Kossiakoff},
doi = {10.1002/pro.3751},
issn = {1469-896X},
year = {2020},
date = {2020-01-01},
urldate = {2020-01-01},
journal = {Protein Sci},
volume = {29},
number = {1},
pages = {141--156},
abstract = {Engineered recombinant antibody-based reagents are rapidly supplanting traditionally derived antibodies in many cell biological applications. A particularly powerful aspect of these engineered reagents is that other modules having myriad functions can be attached to them either chemically or through molecular fusions. However, these processes can be cumbersome and do not lend themselves to high throughput applications. Consequently, we have endeavored to develop a platform that can introduce multiple functionalities into a class of Fab-based affinity reagents in a "plug and play" fashion. This platform exploits the ultra-tight binding interaction between affinity matured variants of a Fab scaffold (Fab ) and a domain of an immunoglobulin binding protein, protein G (GA1). GA1 is easily genetically manipulatable facilitating the ability to link these modules together like beads on a string with adjustable spacing to produce multivalent and bi-specific entities. GA1 can also be fused to other proteins or be chemically modified to engage other types of functional components. To demonstrate the utility for the Fab-GA1 platform, we applied it to a detection proximity assay based on the β-lactamase (BL) split enzyme system. We also show the bi-specific capabilities of the module by using it in context of a Bi-specific T-cell engager (BiTE), which is a therapeutic assemblage that induces cell killing by crosslinking T-cells to cancer cells. We show that GA1-Fab modules are easily engineered into potent cell-killing BiTE-like assemblages and have the advantage of interchanging Fabs directed against different cell surface cancer-related targets in a plug and play fashion.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}