Publications: 2017
2017
Zhang, Wei; Sartori, Maria A; Makhnevych, Taras; Federowicz, Kelly E; Dong, Xiaohui; Liu, Li; Nim, Satra; Dong, Aiping; Yang, Jingsong; Li, Yanjun; Haddad, Dania; Ernst, Andreas; Heerding, Dirk; Tong, Yufeng; Moffat, Jason; Sidhu, Sachdev S
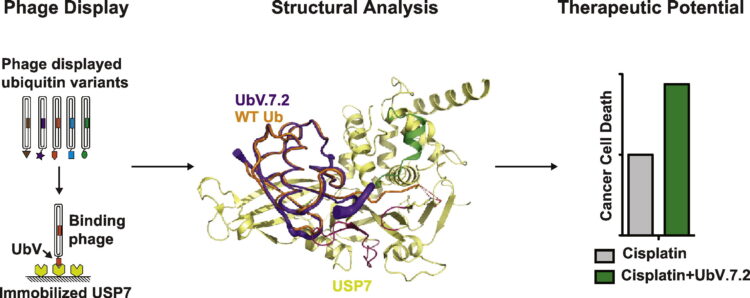
Generation and Validation of Intracellular Ubiquitin Variant Inhibitors for USP7 and USP10 Journal Article
In: J Mol Biol, vol. 429, no. 22, pp. 3546–3560, 2017, ISSN: 1089-8638.
@article{pmid28587923,
title = {Generation and Validation of Intracellular Ubiquitin Variant Inhibitors for USP7 and USP10},
author = {Wei Zhang and Maria A Sartori and Taras Makhnevych and Kelly E Federowicz and Xiaohui Dong and Li Liu and Satra Nim and Aiping Dong and Jingsong Yang and Yanjun Li and Dania Haddad and Andreas Ernst and Dirk Heerding and Yufeng Tong and Jason Moffat and Sachdev S Sidhu},
doi = {10.1016/j.jmb.2017.05.025},
issn = {1089-8638},
year = {2017},
date = {2017-11-01},
urldate = {2017-11-01},
journal = {J Mol Biol},
volume = {429},
number = {22},
pages = {3546--3560},
abstract = {Post-translational modification of the p53 signaling pathway plays an important role in cell cycle progression and stress-induced apoptosis. Indeed, a large body of work has shown that dysregulation of p53 and its E3 ligase MDM2 by the ubiquitin-proteasome system (UPS) promotes carcinogenesis and malignant transformation. Thus, drug discovery efforts have focused on the restoration of wild-type p53 activity or inactivation of oncogenic mutant p53 by targeted inhibition of UPS components, particularly key deubiquitinases (DUBs) of the ubiquitin-specific protease (USP) class. However, development of selective small-molecule USP inhibitors has been challenging, partly due to the highly conserved structural features of the catalytic sites across the class. To tackle this problem, we devised a protein engineering strategy for rational design of inhibitors for DUBs and other UPS proteins. We employed a phage-displayed ubiquitin variant (UbV) library to develop inhibitors targeting the DUBs USP7 and USP10, which are involved in regulating levels of p53 and MDM2. We were able to identify UbVs that bound USP7 or USP10 with high affinity and inhibited deubiquitination activity. We solved the crystal structure of UbV.7.2 and rationalized the molecular basis for enhanced affinity and specificity for USP7. Finally, cell death was increased significantly by UbV.7.2 expression in a colon cancer cell line that was treated with the chemotherapy drug cisplatin, demonstrating the therapeutic potential of inhibiting USP7 by this approach.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Diaz, Juan E; Morgan, Charles W; Minogue, Catherine E; Hebert, Alexander S; Coon, Joshua J; Wells, James A
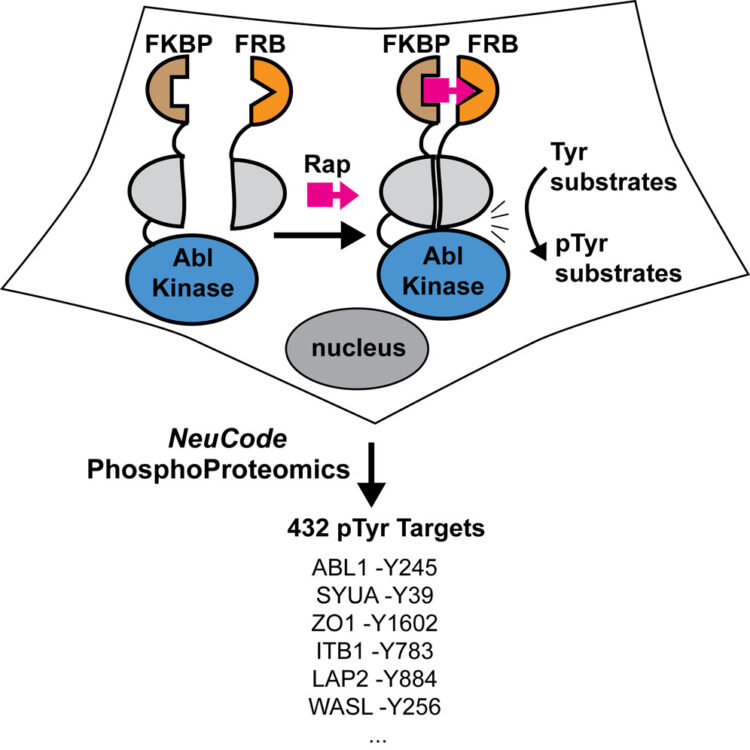
A Split-Abl Kinase for Direct Activation in Cells Journal Article
In: Cell Chem Biol, vol. 24, no. 10, pp. 1250–1258.e4, 2017, ISSN: 2451-9448.
@article{pmid28919041,
title = {A Split-Abl Kinase for Direct Activation in Cells},
author = {Juan E Diaz and Charles W Morgan and Catherine E Minogue and Alexander S Hebert and Joshua J Coon and James A Wells},
doi = {10.1016/j.chembiol.2017.08.007},
issn = {2451-9448},
year = {2017},
date = {2017-10-01},
urldate = {2017-10-01},
journal = {Cell Chem Biol},
volume = {24},
number = {10},
pages = {1250--1258.e4},
abstract = {To dissect the cellular roles of individual kinases, it is useful to design tools for their selective activation. We describe the engineering of a split-cAbl kinase (sKin-Abl) that is rapidly activated in cells with rapamycin and allows temporal, dose, and compartmentalization control. Our design strategy involves an empirical screen in mammalian cells and identification of split site in the N lobe. This split site leads to complete loss of activity, which can be restored upon small-molecule-induced dimerization in cells. Remarkably, the split site is transportable to the related Src Tyr kinase and the distantly related Ser/Thr kinase, AKT, suggesting broader applications to kinases. To quantify the fold induction of phosphotyrosine (pTyr) modification, we employed quantitative proteomics, NeuCode SILAC. We identified a number of known Abl substrates, including autophosphorylation sites and novel pTyr targets, 432 pTyr sites in total. We believe that this split-kinase technology will be useful for direct activation of protein kinases in cells.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Julien, Olivier; Wells, James A
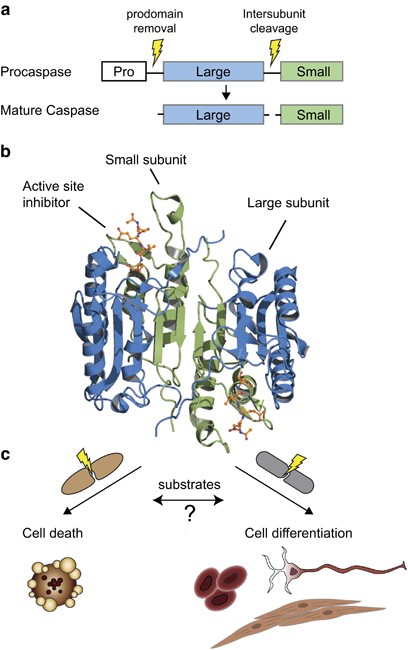
Caspases and their substrates Journal Article
In: Cell Death Differ, vol. 24, no. 8, pp. 1380–1389, 2017, ISSN: 1476-5403.
@article{pmid28498362,
title = {Caspases and their substrates},
author = {Olivier Julien and James A Wells},
doi = {10.1038/cdd.2017.44},
issn = {1476-5403},
year = {2017},
date = {2017-08-01},
urldate = {2017-08-01},
journal = {Cell Death Differ},
volume = {24},
number = {8},
pages = {1380--1389},
abstract = {Protease biology is intimately linked to the functional consequences of substrate cleavage events. Human caspases are a family of 12 fate-determining cysteine proteases that are best known for driving cell death, either apoptosis or pyroptosis. More recently, caspases have been shown to be involved in other cellular remodeling events as well including stem cell fate determination, spermatogenesis, and erythroid differentiation. Recent global proteomics methods enable characterization of the substrates that caspases cleave in live cells and cell extracts. The number of substrate targets identified for individual caspases can vary widely ranging from only a (few) dozen targets for caspases-4, -5, -9, and -14 to hundreds of targets for caspases-1, -2, -3, -6, -7, and -8. Proteomic studies characterizing the rates of target cleavage show that each caspase has a preferred substrate cohort that sometimes overlaps between caspases, but whose rates of cleavage vary over 500-fold within each group. Determining the functional consequences of discrete proteolytic events within the global substrate pool is a major challenge for the field. From the handful of individual targets that have been studied in detail, there are only a few so far that whose single cleavage event is capable of sparking apoptosis alone, such as cleavage of caspase-3/-7 and BIM, or for pyroptosis, gasdermin D. For the most part, it appears that cleavage events function cooperatively in the cell death process to generate a proteolytic synthetic lethal outcome. In contrast to apoptosis, far less is known about caspase biology in non-apoptotic cellular processes, such as cellular remodeling, including which caspases are activated, the mechanisms of their activation and deactivation, and the key substrate targets. Here we survey the progress made in global identification of caspase substrates using proteomics and the exciting new avenues these studies have opened for understanding the molecular logic of substrate cleavage in apoptotic and non-apoptotic processes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
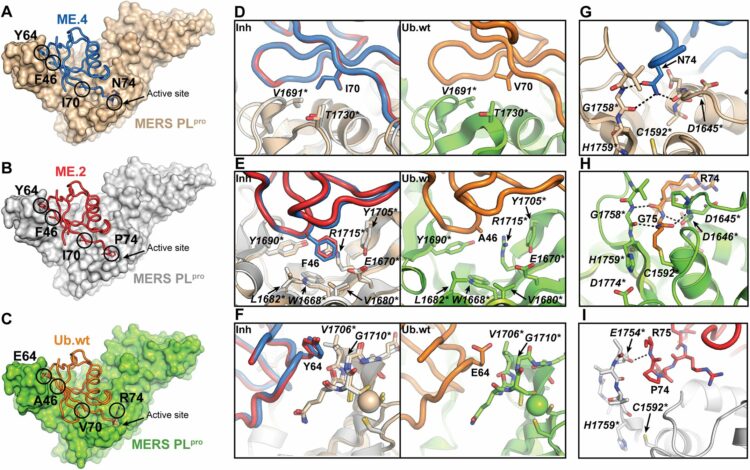
Zhang, Wei; Bailey-Elkin, Ben A; Knaap, Robert C M; Khare, Baldeep; Dalebout, Tim J; Johnson, Garrett G; van Kasteren, Puck B; McLeish, Nigel J; Gu, Jun; He, Wenguang; Kikkert, Marjolein; Mark, Brian L; Sidhu, Sachdev S
Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants Journal Article
In: PLoS Pathog, vol. 13, no. 5, pp. e1006372, 2017, ISSN: 1553-7374.
@article{pmid28542609,
title = {Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants},
author = {Wei Zhang and Ben A Bailey-Elkin and Robert C M Knaap and Baldeep Khare and Tim J Dalebout and Garrett G Johnson and Puck B van Kasteren and Nigel J McLeish and Jun Gu and Wenguang He and Marjolein Kikkert and Brian L Mark and Sachdev S Sidhu},
doi = {10.1371/journal.ppat.1006372},
issn = {1553-7374},
year = {2017},
date = {2017-05-01},
urldate = {2017-05-01},
journal = {PLoS Pathog},
volume = {13},
number = {5},
pages = {e1006372},
abstract = {The recent Middle East respiratory syndrome coronavirus (MERS-CoV), Ebola and Zika virus outbreaks exemplify the continued threat of (re-)emerging viruses to human health, and our inability to rapidly develop effective therapeutic countermeasures. Many viruses, including MERS-CoV and the Crimean-Congo hemorrhagic fever virus (CCHFV) encode deubiquitinating (DUB) enzymes that are critical for viral replication and pathogenicity. They bind and remove ubiquitin (Ub) and interferon stimulated gene 15 (ISG15) from cellular proteins to suppress host antiviral innate immune responses. A variety of viral DUBs (vDUBs), including the MERS-CoV papain-like protease, are responsible for cleaving the viral replicase polyproteins during replication, and are thereby critical components of the viral replication cycle. Together, this makes vDUBs highly attractive antiviral drug targets. However, structural similarity between the catalytic cores of vDUBs and human DUBs complicates the development of selective small molecule vDUB inhibitors. We have thus developed an alternative strategy to target the vDUB activity through a rational protein design approach. Here, we report the use of phage-displayed ubiquitin variant (UbV) libraries to rapidly identify potent and highly selective protein-based inhibitors targeting the DUB domains of MERS-CoV and CCHFV. UbVs bound the vDUBs with high affinity and specificity to inhibit deubiquitination, deISGylation and in the case of MERS-CoV also viral replicative polyprotein processing. Co-crystallization studies further revealed critical molecular interactions between UbVs and MERS-CoV or CCHFV vDUBs, accounting for the observed binding specificity and high affinity. Finally, expression of UbVs during MERS-CoV infection reduced infectious progeny titers by more than four orders of magnitude, demonstrating the remarkable potency of UbVs as antiviral agents. Our results thereby establish a strategy to produce protein-based inhibitors that could protect against a diverse range of viruses by providing UbVs via mRNA or protein delivery technologies or through transgenic techniques.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
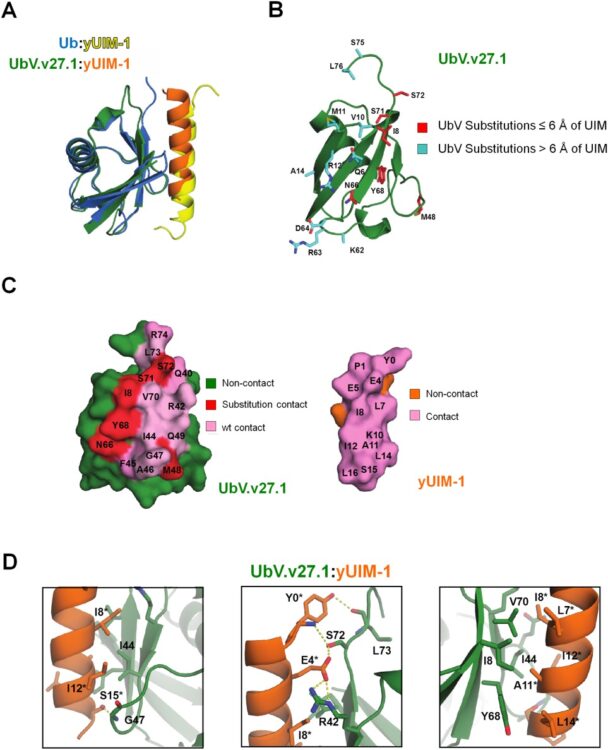
Manczyk, Noah; Yates, Bradley P; Veggiani, Gianluca; Ernst, Andreas; Sicheri, Frank; Sidhu, Sachdev S
In: Protein Sci, vol. 26, no. 5, pp. 1060–1069, 2017, ISSN: 1469-896X.
@article{pmid28276594,
title = {Structural and functional characterization of a ubiquitin variant engineered for tight and specific binding to an alpha-helical ubiquitin interacting motif},
author = {Noah Manczyk and Bradley P Yates and Gianluca Veggiani and Andreas Ernst and Frank Sicheri and Sachdev S Sidhu},
doi = {10.1002/pro.3155},
issn = {1469-896X},
year = {2017},
date = {2017-05-01},
urldate = {2017-05-01},
journal = {Protein Sci},
volume = {26},
number = {5},
pages = {1060--1069},
abstract = {Ubiquitin interacting motifs (UIMs) are short α-helices found in a number of eukaryotic proteins. UIMs interact weakly but specifically with ubiquitin conjugated to other proteins, and in so doing, mediate specific cellular signals. Here we used phage display to generate ubiquitin variants (UbVs) targeting the N-terminal UIM of the yeast Vps27 protein. Selections yielded UbV.v27.1, which recognized the cognate UIM with high specificity relative to other yeast UIMs and bound with an affinity more than two orders of magnitude higher than that of ubiquitin. Structural and mutational studies of the UbV.v27.1-UIM complex revealed the molecular details for the enhanced affinity and specificity of UbV.v27.1, and underscored the importance of changes at the binding interface as well as at positions that do not contact the UIM. Our study highlights the power of the phage display approach for selecting UbVs with unprecedented affinity and high selectivity for particular α-helical UIM domains within proteomes, and it establishes a general approach for the development of inhibitors targeting interactions of this type.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
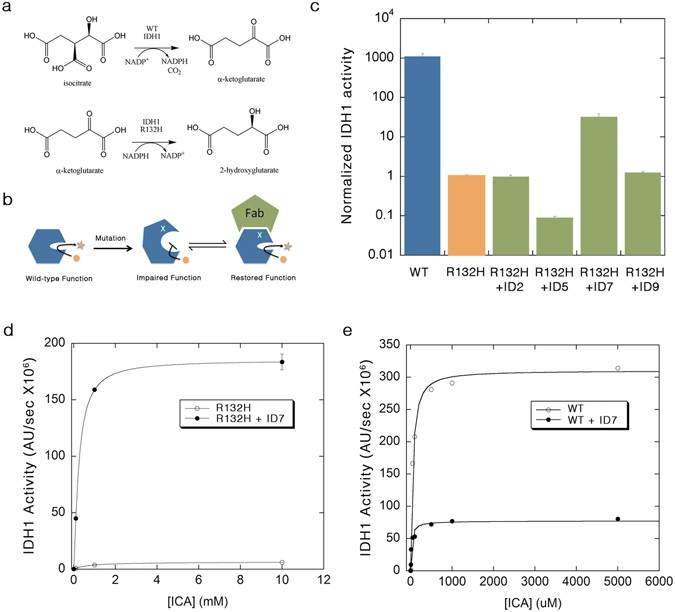
Rizk, Shahir S; Mukherjee, Somnath; Koide, Akiko; Koide, Shohei; Kossiakoff, Anthony A
Targeted rescue of cancer-associated IDH1 mutant activity using an engineered synthetic antibody Journal Article
In: Sci Rep, vol. 7, no. 1, pp. 556, 2017, ISSN: 2045-2322.
@article{pmid28373671,
title = {Targeted rescue of cancer-associated IDH1 mutant activity using an engineered synthetic antibody},
author = {Shahir S Rizk and Somnath Mukherjee and Akiko Koide and Shohei Koide and Anthony A Kossiakoff},
doi = {10.1038/s41598-017-00728-1},
issn = {2045-2322},
year = {2017},
date = {2017-04-01},
urldate = {2017-04-01},
journal = {Sci Rep},
volume = {7},
number = {1},
pages = {556},
abstract = {We have utilized a high-diversity phage display library to engineer antibody fragments (Fabs) that can modulate the activity of the enzyme isocitrate dehydrogenase 1 (IDH1). We show that a conformation-specific Fab can reactivate an IDH1 mutant associated with brain tumors. The results show that this strategy is a first step towards developing "activator drugs" for a large number of genetic disorders where mutations have disrupted protein function.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
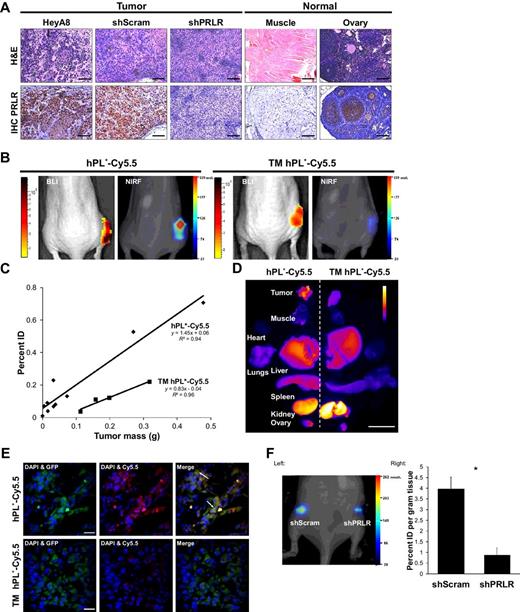
Sundaram, Karthik M; Zhang, Yilin; Mitra, Anirban K; Kouadio, Jean-Louis K; Gwin, Katja; Kossiakoff, Anthony A; Roman, Brian B; Lengyel, Ernst; Piccirilli, Joseph A
Prolactin Receptor-Mediated Internalization of Imaging Agents Detects Epithelial Ovarian Cancer with Enhanced Sensitivity and Specificity Journal Article
In: Cancer Res, vol. 77, no. 7, pp. 1684–1696, 2017, ISSN: 1538-7445.
@article{pmid28202518,
title = {Prolactin Receptor-Mediated Internalization of Imaging Agents Detects Epithelial Ovarian Cancer with Enhanced Sensitivity and Specificity},
author = {Karthik M Sundaram and Yilin Zhang and Anirban K Mitra and Jean-Louis K Kouadio and Katja Gwin and Anthony A Kossiakoff and Brian B Roman and Ernst Lengyel and Joseph A Piccirilli},
doi = {10.1158/0008-5472.CAN-16-1454},
issn = {1538-7445},
year = {2017},
date = {2017-04-01},
urldate = {2017-04-01},
journal = {Cancer Res},
volume = {77},
number = {7},
pages = {1684--1696},
abstract = {Poor prognosis of ovarian cancer, the deadliest of the gynecologic malignancies, reflects major limitations associated with detection and diagnosis. Current methods lack high sensitivity to detect small tumors and high specificity to distinguish malignant from benign tissue, both impeding diagnosis of early and metastatic cancer stages and leading to costly and invasive surgeries. Tissue microarray analysis revealed that >98% of ovarian cancers express the prolactin receptor (PRLR), forming the basis of a new molecular imaging strategy. We fused human placental lactogen (hPL), a specific and tight binding PRLR ligand, to magnetic resonance imaging (gadolinium) and near-infrared fluorescence imaging agents. Both in tissue culture and in mouse models, these imaging bioconjugates underwent selective internalization into ovarian cancer cells via PRLR-mediated endocytosis. Compared with current clinical MRI techniques, this targeted approach yielded both enhanced signal-to-noise ratio from accumulation of signal via selective internalization and improved specificity conferred by PRLR upregulation in malignant ovarian cancer. These features endow PRLR-targeted imaging with the potential to transform ovarian cancer detection. .},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Calvo, Sarah E; Julien, Olivier; Clauser, Karl R; Shen, Hongying; Kamer, Kimberli J; Wells, James A; Mootha, Vamsi K
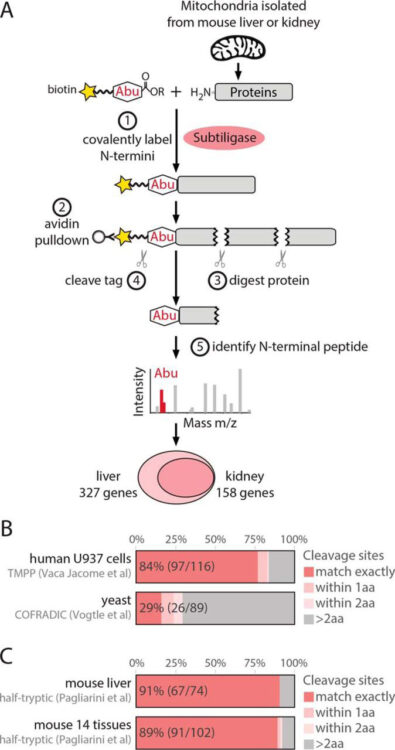
Comparative Analysis of Mitochondrial N-Termini from Mouse, Human, and Yeast Journal Article
In: Mol Cell Proteomics, vol. 16, no. 4, pp. 512–523, 2017, ISSN: 1535-9484.
@article{pmid28122942b,
title = {Comparative Analysis of Mitochondrial N-Termini from Mouse, Human, and Yeast},
author = {Sarah E Calvo and Olivier Julien and Karl R Clauser and Hongying Shen and Kimberli J Kamer and James A Wells and Vamsi K Mootha},
doi = {10.1074/mcp.M116.063818},
issn = {1535-9484},
year = {2017},
date = {2017-04-01},
urldate = {2017-04-01},
journal = {Mol Cell Proteomics},
volume = {16},
number = {4},
pages = {512--523},
abstract = {The majority of mitochondrial proteins are encoded in the nuclear genome, translated in the cytoplasm, and directed to the mitochondria by an N-terminal presequence that is cleaved upon import. Recently, N-proteome catalogs have been generated for mitochondria from yeast and from human U937 cells. Here, we applied the subtiligase method to determine N-termini for 327 proteins in mitochondria isolated from mouse liver and kidney. Comparative analysis between mitochondrial N-termini from mouse, human, and yeast proteins shows that whereas presequences are poorly conserved at the sequence level, other presequence properties are extremely conserved, including a length of ∼20-60 amino acids, a net charge between +3 to +6, and the presence of stabilizing amino acids at the N-terminus of mature proteins that follow the N-end rule from bacteria. As in yeast, ∼80% of mouse presequence cleavage sites match canonical motifs for three mitochondrial peptidases (MPP, Icp55, and Oct1), whereas the remainder do not match any known peptidase motifs. We show that mature mitochondrial proteins often exist with a spectrum of N-termini, consistent with a model of multiple cleavage events by MPP and Icp55. In addition to analysis of canonical targeting presequences, our N-terminal dataset allows the exploration of other cleavage events and provides support for polypeptide cleavage into two distinct enzymes (Hsd17b4), protein cleavages key for signaling (Oma1, Opa1, Htra2, Mavs, and Bcs2l13), and in several cases suggests novel protein isoforms (Scp2, Acadm, Adck3, Hsdl2, Dlst, and Ogdh). We present an integrated catalog of mammalian mitochondrial N-termini that can be used as a community resource to investigate individual proteins, to elucidate mechanisms of mammalian mitochondrial processing, and to allow researchers to engineer tags distally to the presequence cleavage.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
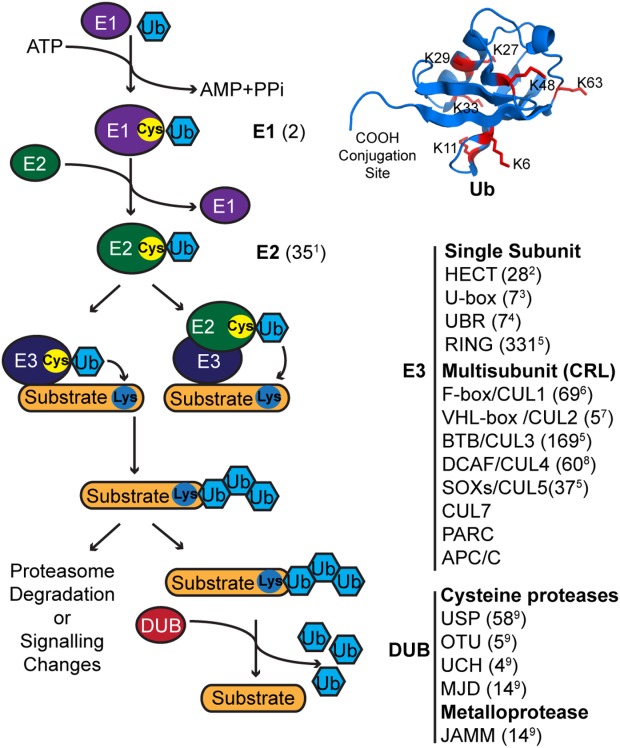
Gorelik, Maryna; Sidhu, Sachdev S
Specific targeting of the deubiquitinase and E3 ligase families with engineered ubiquitin variants Journal Article
In: Bioeng Transl Med, vol. 2, no. 1, pp. 31–42, 2017, ISSN: 2380-6761.
@article{pmid28580429,
title = {Specific targeting of the deubiquitinase and E3 ligase families with engineered ubiquitin variants},
author = {Maryna Gorelik and Sachdev S Sidhu},
doi = {10.1002/btm2.10044},
issn = {2380-6761},
year = {2017},
date = {2017-03-01},
urldate = {2017-03-01},
journal = {Bioeng Transl Med},
volume = {2},
number = {1},
pages = {31--42},
abstract = {The ubiquitin proteasome system (UPS) has garnered much attention due to its potential for the development of therapeutics. Following a successful clinical application of general proteasome inhibitors much effort has been devoted to targeting individual UPS components including E3 enzymes and deubiquitinases that control specificity of ubiquitination. Our group has developed a novel approach for targeting the UPS proteins using engineered ubiquitin variants (Ubvs). These drug-like proteins can serve as valuable tools to study biological function of UPS components and assist in the development of small molecules for clinical use. In this review, we summarize studies of Ubvs targeting members of three major families, including deubiquitinases, HECT E3 ligases, and CRL E3 ligases. In particular, we focus on Ubv binding mechanisms, structural studies, and effects on enzyme function. Furthermore, new insights gained from the Ubvs are discussed in the context of small molecule studies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
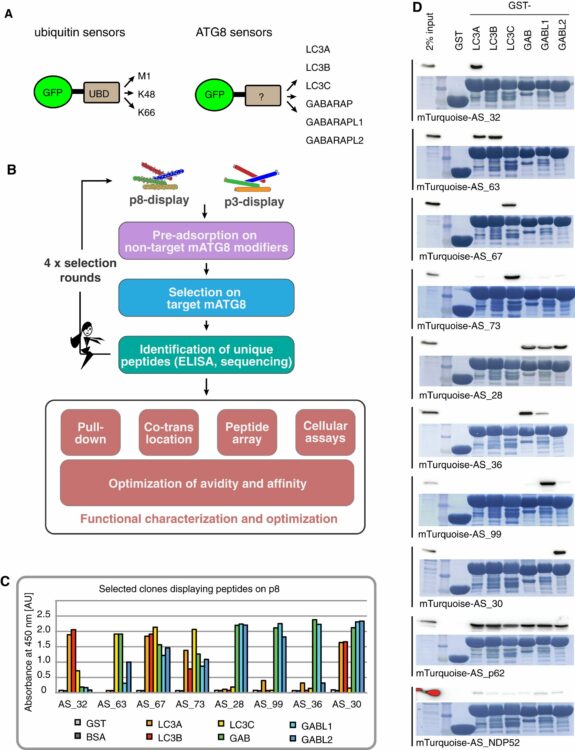
Stolz, Alexandra; Putyrski, Mateusz; Kutle, Ivana; Huber, Jessica; Wang, Chunxin; Major, Viktória; Sidhu, Sachdev S; Youle, Richard J; Rogov, Vladimir V; Dötsch, Volker; Ernst, Andreas; Dikic, Ivan
Fluorescence-based ATG8 sensors monitor localization and function of LC3/GABARAP proteins Journal Article
In: EMBO J, vol. 36, no. 4, pp. 549–564, 2017, ISSN: 1460-2075.
@article{pmid28028054,
title = {Fluorescence-based ATG8 sensors monitor localization and function of LC3/GABARAP proteins},
author = {Alexandra Stolz and Mateusz Putyrski and Ivana Kutle and Jessica Huber and Chunxin Wang and Viktória Major and Sachdev S Sidhu and Richard J Youle and Vladimir V Rogov and Volker Dötsch and Andreas Ernst and Ivan Dikic},
doi = {10.15252/embj.201695063},
issn = {1460-2075},
year = {2017},
date = {2017-02-01},
urldate = {2017-02-01},
journal = {EMBO J},
volume = {36},
number = {4},
pages = {549--564},
abstract = {Autophagy is a cellular surveillance pathway that balances metabolic and energy resources and transports specific cargos, including damaged mitochondria, other broken organelles, or pathogens for degradation to the lysosome. Central components of autophagosomal biogenesis are six members of the LC3 and GABARAP family of ubiquitin-like proteins (mATG8s). We used phage display to isolate peptides that possess LIR (LC3-interacting region) properties and are selective for individual mATG8 isoforms. Sensitivity of the developed sensors was optimized by multiplication, charge distribution, and fusion with a membrane recruitment (FYVE) or an oligomerization (PB1) domain. We demonstrate the use of the engineered peptides as intracellular sensors that recognize specifically GABARAP, GABL1, GABL2, and LC3C, as well as a bispecific sensor for LC3A and LC3B. By using an LC3C-specific sensor, we were able to monitor recruitment of endogenous LC3C to during xenophagy, as well as to mitochondria during mitophagy. The sensors are general tools to monitor the fate of mATG8s and will be valuable in decoding the biological functions of the individual LC3/GABARAPs.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
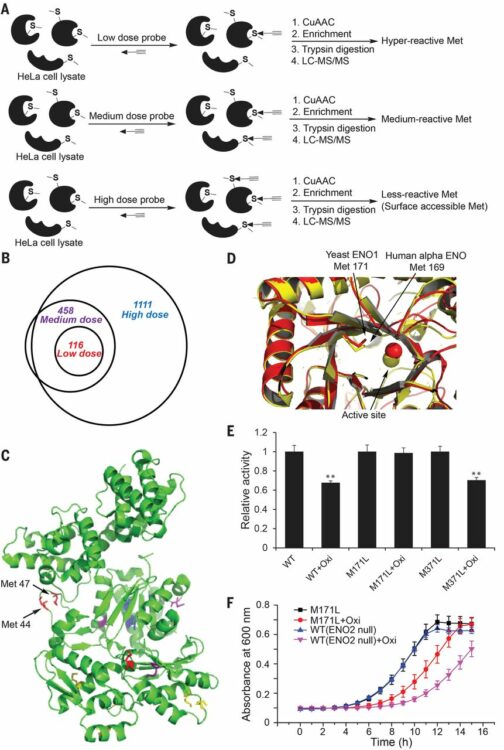
Lin, Shixian; Yang, Xiaoyu; Jia, Shang; Weeks, Amy M; Hornsby, Michael; Lee, Peter S; Nichiporuk, Rita V; Iavarone, Anthony T; Wells, James A; Toste, F Dean; Chang, Christopher J
Redox-based reagents for chemoselective methionine bioconjugation Journal Article
In: Science, vol. 355, no. 6325, pp. 597–602, 2017, ISSN: 1095-9203.
@article{pmid28183972,
title = {Redox-based reagents for chemoselective methionine bioconjugation},
author = {Shixian Lin and Xiaoyu Yang and Shang Jia and Amy M Weeks and Michael Hornsby and Peter S Lee and Rita V Nichiporuk and Anthony T Iavarone and James A Wells and F Dean Toste and Christopher J Chang},
doi = {10.1126/science.aal3316},
issn = {1095-9203},
year = {2017},
date = {2017-02-01},
urldate = {2017-02-01},
journal = {Science},
volume = {355},
number = {6325},
pages = {597--602},
abstract = {Cysteine can be specifically functionalized by a myriad of acid-base conjugation strategies for applications ranging from probing protein function to antibody-drug conjugates and proteomics. In contrast, selective ligation to the other sulfur-containing amino acid, methionine, has been precluded by its intrinsically weaker nucleophilicity. Here, we report a strategy for chemoselective methionine bioconjugation through redox reactivity, using oxaziridine-based reagents to achieve highly selective, rapid, and robust methionine labeling under a range of biocompatible reaction conditions. We highlight the broad utility of this conjugation method to enable precise addition of payloads to proteins, synthesis of antibody-drug conjugates, and identification of hyperreactive methionine residues in whole proteomes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Chen, Gang; Sidhu, Sachdev S; Nilvebrant, Johan
Synthetic Antibodies in Infectious Disease Journal Article
In: Adv Exp Med Biol, vol. 1053, pp. 79–98, 2017, ISSN: 0065-2598.
@article{pmid29549636,
title = {Synthetic Antibodies in Infectious Disease},
author = {Gang Chen and Sachdev S Sidhu and Johan Nilvebrant},
doi = {10.1007/978-3-319-72077-7_5},
issn = {0065-2598},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
journal = {Adv Exp Med Biol},
volume = {1053},
pages = {79--98},
abstract = {Rapid spread of microbial resistance and recent outbreaks of viral disease have led to renewed interest in antibody-based therapies for infectious diseases. Synthetic antibody libraries displayed on phage offer unique advantages over traditional immunization-based antibody generation, including full control over library design and selection conditions. The technology has matured beyond natural antibodies and is capable of providing novel insights into infectious disease and can generate novel antibodies that cannot be produced by the natural immune system. This chapter gives an overview of recombinant antibody library technology with an emphasis on our work using a highly successful synthetic single framework Fab library. We demonstrate its utility in targeting viruses and bacterial toxins in five case studies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
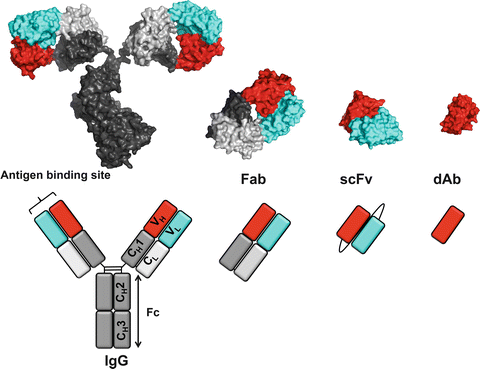
Paduch, Marcin; Kossiakoff, Anthony A
Generating Conformation and Complex-Specific Synthetic Antibodies Journal Article
In: Methods Mol Biol, vol. 1575, pp. 93–119, 2017, ISSN: 1940-6029.
@article{pmid28255876,
title = {Generating Conformation and Complex-Specific Synthetic Antibodies},
author = {Marcin Paduch and Anthony A Kossiakoff},
doi = {10.1007/978-1-4939-6857-2_6},
issn = {1940-6029},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
journal = {Methods Mol Biol},
volume = {1575},
pages = {93--119},
abstract = {Phage display is commonly used to identify and isolate binders from large combinatorial libraries. Here we present phage selection protocols enabling generation of synthetic antibodies capable of recognizing multiprotein complexes and conformational states. The procedure describes stages of the experiment design, optimization, and screening, as well as provides the framework for building downstream assays with an end goal of isolating bioactive antibodies for future therapeutic use. The methods described are also applicable to screening directly on cells and can be ported to other in vitro directed evolution systems utilizing non-immunoglobulin scaffolds.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
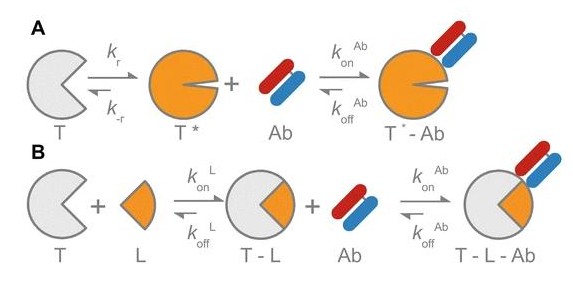
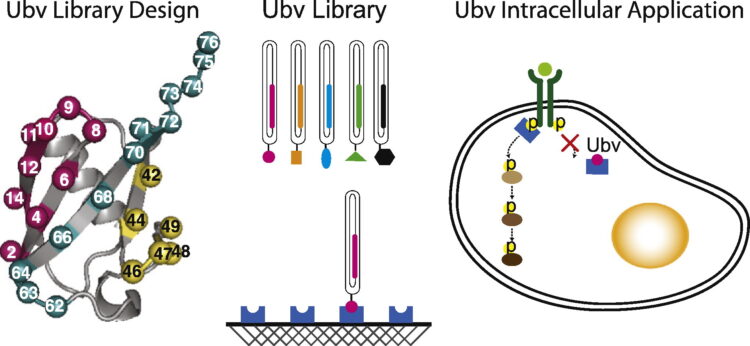
Leung, Isabel; Jarvik, Nick; Sidhu, Sachdev S
A Highly Diverse and Functional Naïve Ubiquitin Variant Library for Generation of Intracellular Affinity Reagents Journal Article
In: J Mol Biol, vol. 429, no. 1, pp. 115–127, 2017, ISSN: 1089-8638.
@article{pmid27887869,
title = {A Highly Diverse and Functional Naïve Ubiquitin Variant Library for Generation of Intracellular Affinity Reagents},
author = {Isabel Leung and Nick Jarvik and Sachdev S Sidhu},
doi = {10.1016/j.jmb.2016.11.016},
issn = {1089-8638},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
journal = {J Mol Biol},
volume = {429},
number = {1},
pages = {115--127},
abstract = {We report the design, construction, and validation of a highly diverse phage-displayed naïve ubiquitin variant (Ubv) library. We first conducted a mutation tolerance scan of 27 residues and confirmed that 24 of these could be substituted by chemically diverse amino acids without compromising the display of Ubvs on phage. Subsequently, we constructed a library containing 6.8×10 unique members, in which these 24 positions were diversified with a degenerate codon that encodes for 6 aa that are prevalent in protein interaction sites. To ensure the optimal structural stability of the Ubvs, we constructed the library in a two-step process, whereby 12 positions were randomized first, and following the selection for displayed Ubvs, the resulting pool was further diversified at the other 12 positions. The resulting library was validated by conducting binding selections against a panel of 40 diverse protein antigens and was found to be as functional as a highly validated synthetic antibody library, yielding binders against 30 of the antigens. Detailed characterization of an Ubv that bound to the cell-surface receptor human epidermal growth factor receptor 3 revealed tight binding in the single-digit nanomolar range. Moreover, Ubvs that bound to two distinct sites on the intracellular adapter Grb2 could be combined to generate a potent inhibitor that functioned in cells. These results validate ubiquitin as a robust scaffold for the construction of naïve libraries that can be used to generate Ubvs that target signaling networks both outside and inside the cells.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
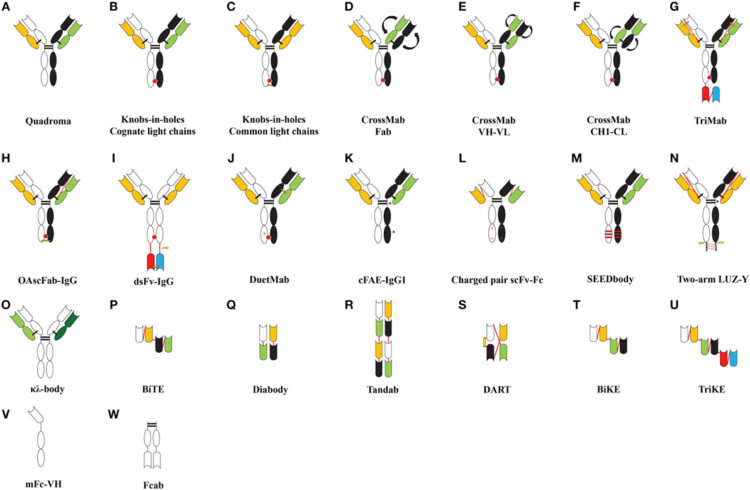
Liu, Hongyan; Saxena, Abhishek; Sidhu, Sachdev S; Wu, Donghui
Fc Engineering for Developing Therapeutic Bispecific Antibodies and Novel Scaffolds Journal Article
In: Front Immunol, vol. 8, pp. 38, 2017, ISSN: 1664-3224.
@article{pmid28184223,
title = {Fc Engineering for Developing Therapeutic Bispecific Antibodies and Novel Scaffolds},
author = {Hongyan Liu and Abhishek Saxena and Sachdev S Sidhu and Donghui Wu},
doi = {10.3389/fimmu.2017.00038},
issn = {1664-3224},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
journal = {Front Immunol},
volume = {8},
pages = {38},
abstract = {Therapeutic monoclonal antibodies have become molecules of choice to treat autoimmune disorders, inflammatory diseases, and cancer. Moreover, bispecific/multispecific antibodies that target more than one antigen or epitope on a target cell or recruit effector cells (T cell, natural killer cell, or macrophage cell) toward target cells have shown great potential to maximize the benefits of antibody therapy. In the past decade, many novel concepts to generate bispecific and multispecific antibodies have evolved successfully into a range of formats from full bispecific immunoglobulin gammas to antibody fragments. Impressively, antibody fragments such as bispecific T-cell engager, bispecific killer cell engager, trispecific killer cell engager, tandem diabody, and dual-affinity-retargeting are showing exciting results in terms of recruiting and activating self-immune effector cells to target and lyse tumor cells. Promisingly, crystallizable fragment (Fc) antigen-binding fragment and monomeric antibody or half antibody may be particularly advantageous to target solid tumors owing to their small size and thus good tissue penetration potential while, on the other hand, keeping Fc-related effector functions such as antibody-dependent cellular cytotoxicity, complement-dependent cytotoxicity, antibody-dependent cell-mediated phagocytosis, and extended serum half-life interaction with neonatal Fc receptor. This review, therefore, focuses on the progress of Fc engineering in generating bispecific molecules and on the use of small antibody fragment as scaffolds for therapeutic development.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
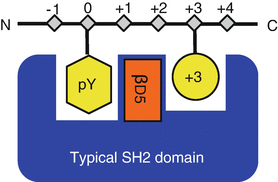
Huang, Haiming; Kaneko, Tomonori; Sidhu, Sachdev S; Li, Shawn S C
Creation of Phosphotyrosine Superbinders by Directed Evolution of an SH2 Domain Journal Article
In: Methods Mol Biol, vol. 1555, pp. 225–254, 2017, ISSN: 1940-6029.
@article{pmid28092036,
title = {Creation of Phosphotyrosine Superbinders by Directed Evolution of an SH2 Domain},
author = {Haiming Huang and Tomonori Kaneko and Sachdev S Sidhu and Shawn S C Li},
doi = {10.1007/978-1-4939-6762-9_13},
issn = {1940-6029},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
journal = {Methods Mol Biol},
volume = {1555},
pages = {225--254},
abstract = {Commercial antibodies raised against phosphotyrosine have been widely used as reagents to detect or isolate tyrosine-phosphorylated proteins from cellular samples. However, these antibodies are costly and are not amenable to in-house production in an academic lab setting. In this chapter, we describe a method to generate super-high affinity SH2 domains, dubbed the phosphotyrosine superbinders, by evolving a natural SH2 domain using the phage display technology. The superbinders are stable and can be easily produced in Escherichia coli in large quantities. The strategy presented here may also be applied to other protein domains to generate domain variants with markedly enhanced affinities for a specific post-translational modification.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}