Technologies
Exploiting the power of phage display technologies.
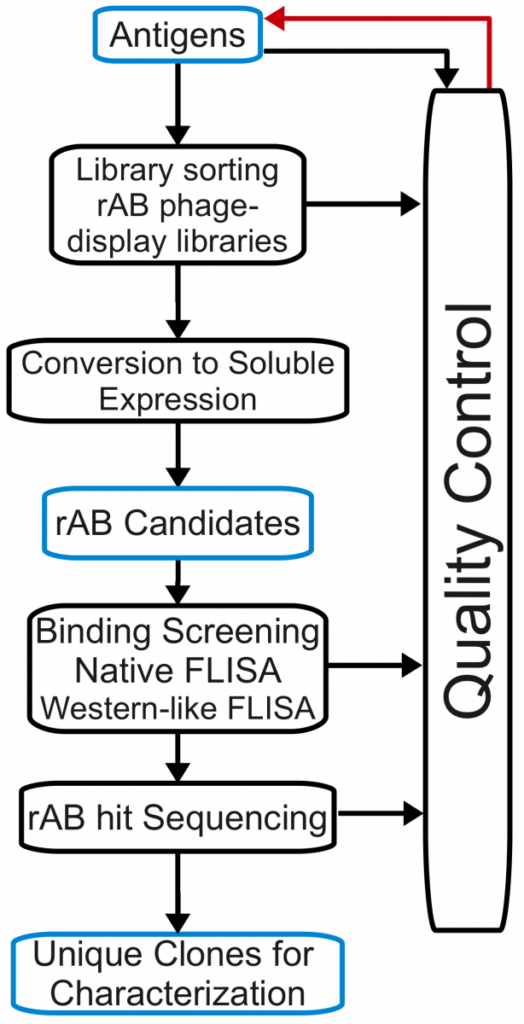
We employ highly tuned phage display selection strategies for producing next generation high performance affinity reagents.
Over the past decade, the accumulation of detailed knowledge of antibody structure and function has enabled phage-displayed antibody libraries to emerge as uniquely powerful in vitro methods.
In phage display, libraries of antibody fragments are usually expressed on the phage surface as single copy fusions to coat proteins.
Phage pools containing billions of unique antibodies can be used in selections to isolate antibodies that recognize antigens of interest, and the sequence of each antibody can be decoded by sequencing of viral DNA.
Thus, phage display not only yields functional antibodies, but also DNA sequences that can be used to produce the protein directly from bacteria and (by mutation) to rapidly generate antibody variants.
Since selections are performed in vitro, the conditions can be tailored to suit the demands of the antigen and intended applications. These features obviate many of the limitations of hybridoma methodology.
Lastly, because the entire process relies mostly on liquid handling and scalable molecular biology techniques, phage display is also suitable for HTP applications.

University of Chicago Robotics
University of Toronto Robotics
University of California at San Francisco Robotics
Exploiting the power of phage display technologies.
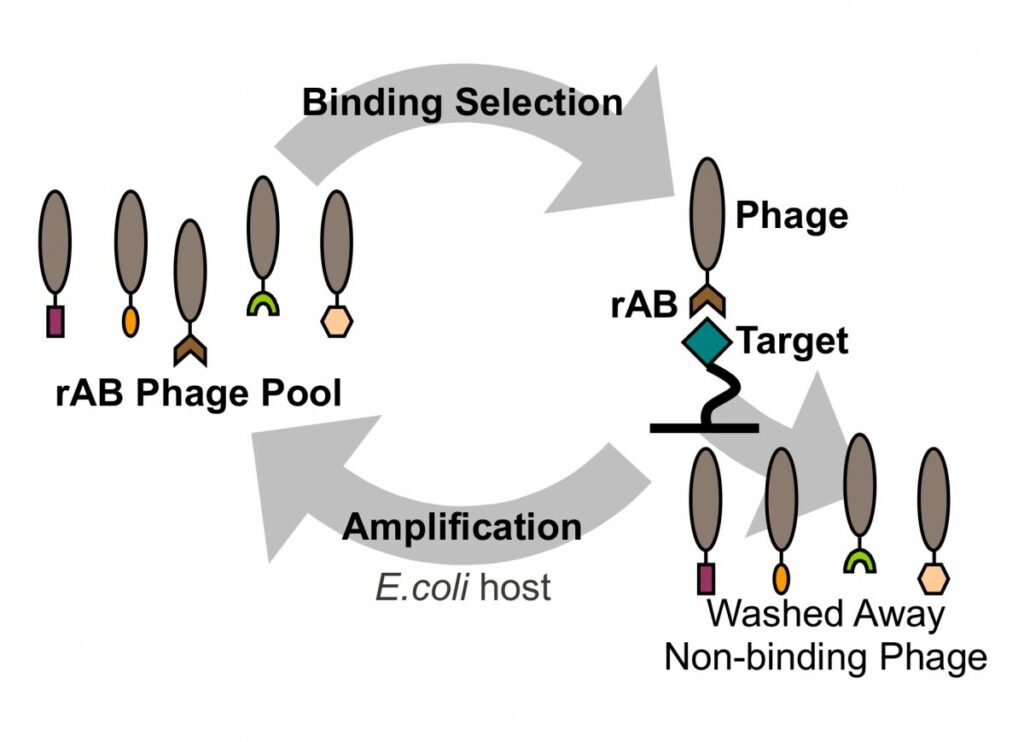
The phage display selection process lends itself extremely well to HTP automation.
It is possible to perform selections on antigens in a 96 well format. So for a particular run, the user has considerable flexibility to either aim for capacity; that is, many targets under one set of selection conditions or fewer targets with several selection conditions.
The latter scenario has proven valuable for the selection of reagents that perform specific functions like capturing particular conformational states or binding to a particular region on the antigen surface. Generally it takes 3-4 rounds of selection to isolate a rich cohort of antibody binders.
This takes approximately a week, but using the automation that the RAN groups will have in place, the process can be staged such that multiple selections can be marched through the pipeline at the same time. Literally hundreds of targets can be processed in a month.
Clearly, the potential of this type of throughput would completely overwhelm the downstream validation part of the pipeline; however, the extraordinary capacity of phage display selection to generate a multitude of binders to a protein target provides an extensive pool of candidates from which the best can be rapidly identified and carried forward for subsequent validation.
Novel Synthetic antibody libraries based on a reduced genetic code.
A particularly powerful advance of our phage display approach is the development of “synthetic” antibody libraries with antigen-binding sites constructed entirely from man-made diversity. Most phage-displayed repertoires are “natural” antibody libraries derived by transferring natural antibody genes to phage display vectors. In contrast, synthetic antibody libraries are created by introducing degenerate, synthetic DNA into the regions encoding the complementarity-determining regions (CDRs) that make up the antigen-binding site within defined variable domain frameworks.
The construction of such libraries requires detailed knowledge of antibody structure and function, as attempts to introduce diversity can lead to protein misfolding if inappropriate chemical diversity or structural sites are used. Despite these difficulties, careful consideration of such issues has led to development of highly functional synthetic antibody libraries that rival or exceed the recognition potential of natural immune repertoires. Numerous examples shown in the preliminary data show that synthetic libraries are more specific than natural libraries.
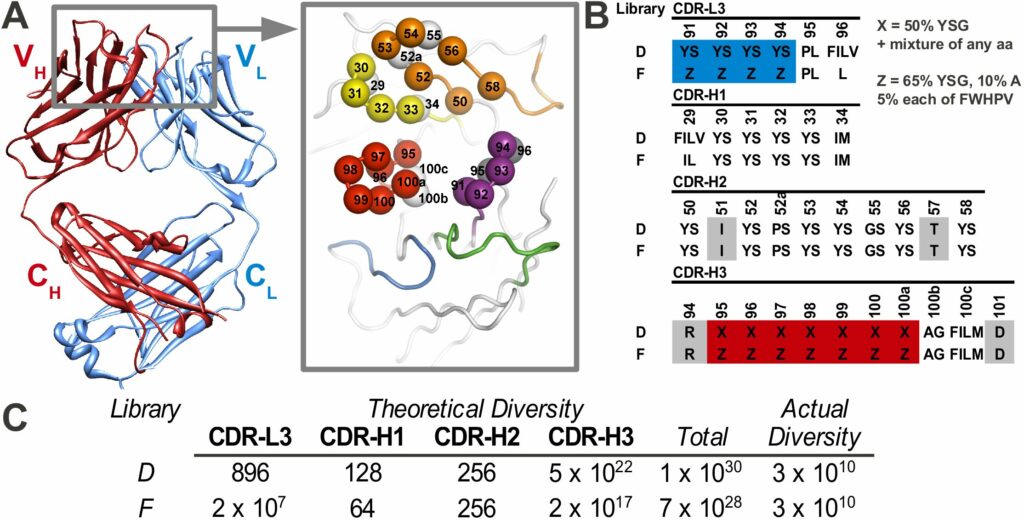
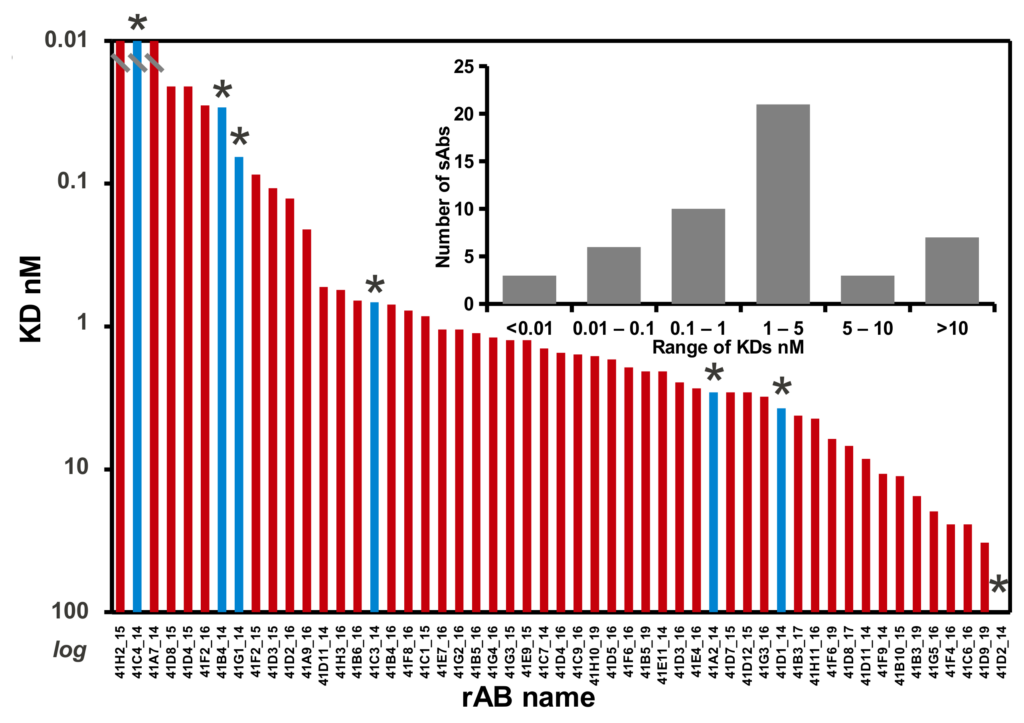
The Toronto and Chicago libraries are unique in that they use a reduced genetic code strategy that has been shown to be both simple and powerful. In fact, they generally produce rABs with nM affinities without needing further affinity maturation.
Synthetic antibodies provide many additional advantages over natural repertoires. Natural repertoires are limited in antibodies against self-antigens, but synthetic antibodies are constructed entirely in vitro and thus are not biased against natural proteins, regardless of source or sequence. Frameworks for synthetic recombinant antibodies can be chosen for particular properties including, for example, high stability and expression. For therapeutic applications, optimized human frameworks can be used to minimize the risk of immunogenicity, thus obviating the need for humanization.
Furthermore, design features can be incorporated to allow facile affinity maturation and adaptation to a HTP pipeline. We will use both synthetic antibody libraries and naturally occurring naïve antibody libraries as sources of rAbs to ensure maximal coverage of any target we study.
Advantages of Fab frameworks.
The Fab framework used by the RAN is based on a very stable antibody fragment. Fabs are robust, easily expressed and very amenable to phage display manipulations. They can be cheaply and efficiently purified by well established protocols.
We refer to the Fab-based affinity binders used in our research as “recombinant antibodies” or rABs.
They have a long history as selective and high affinity capture reagents. rABs can be engineered to introduce fluorescent tags (detectors) or biotin tags for pulldowns (handles).
They can be used as is or oligomerized to exploit increased avidity effects. rABs can be easily reformatted into full length IgG antibodies.
The IgG format has numerous advantages as the format for proteome binders, leveraging a century of scientific effort to develop a diverse menu of technologies that work in many kinds of experiments such as ELISA, Western blots, flow cytometry, bead coupling, immunohistochemistry (IHC) and immunofluorescence (IF). The IgG format is extremely well studied, and is the best characterized and understood of potential proteome binders.
The flexibility of IgGs comes from their modular domain structure. The bivalent binding sites of an IgG enhance affinity through avidity effects and the Fc portion provides a common detection handle.
The IgG format is compatible with current experimental systems and infrastructure used by biochemists, pathologists, and cellular and molecular biologists and all the required secondary reagents are available.
Multifunctional capability of rABs.
In vitro phage display selections are run under highly controlled conditions allowing for generation of rABs that can be “evolved” for specific functions.
For instance, it is readily possible to select for rABs that can: 1) target specific regions on the surface of a protein, 2) recognize specific conformational or oligomeric states, 3) induce conformational changes, 4) capture and stabilize multi-protein complexes, and 5) discriminate between protein isoforms.
These powerful capabilities open a multitude of opportunities for studying complex macromolecular processes at a level inaccessible to current technology. These attributes are particularly relevant to the task of generating “ChIP-able” rAB affinity reagents to the human TFs. TFs are flexible multidomain proteins that generally function bound to DNA and in the context of larger macromolecular complexes. Since the plan is to express two small compact domains of the individual TFs as the antigen targets (one proposed to be the DNA binding domain), it will be important to generate a family of rABs that bind to different epitopes and conformations because it will not be known a priori what will be accessible to the rAB in the assay environment.
Further, it is known that the DNA binding domains of TFs can undergo large conformational changes on binding DNA. Since in many cases the cognate DNA for these domains will not be known, selections will be done using their apo- forms. Thus, we will have to generate a wide spectrum of rABs to ensure that our pool will contain candidates that will work in ChIP experiments.
